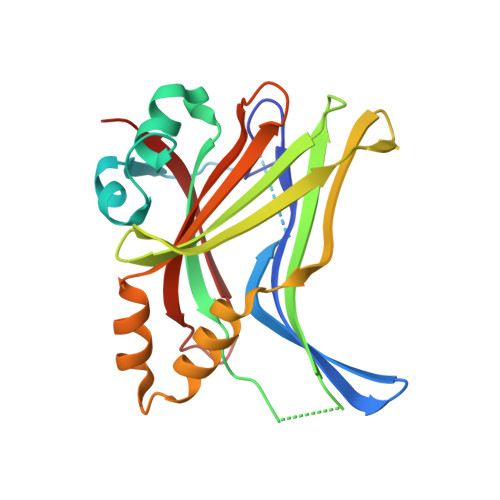
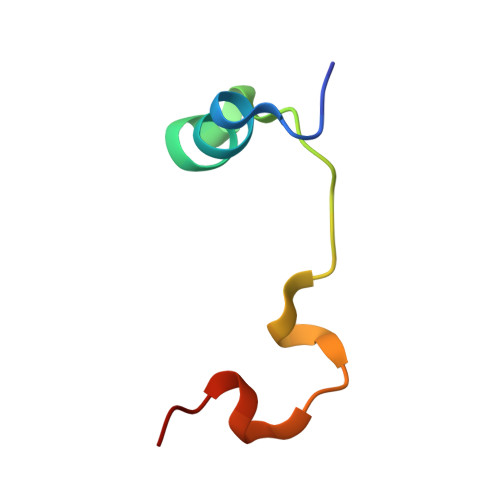
Adaptation of the bound intrinsically disordered protein YAP to mutations at the YAP:TEAD interface.
Mesrouze, Y., Bokhovchuk, F., Izaac, A., Meyerhofer, M., Zimmermann, C., Fontana, P., Schmelzle, T., Erdmann, D., Furet, P., Kallen, J., Chene, P.(2018) Protein Sci 27: 1810-1820
- PubMed: 30058229
- DOI: https://doi.org/10.1002/pro.3493
- Primary Citation of Related Structures:
6GE3, 6GE4, 6GE5, 6GE6, 6GEC, 6GEE, 6GEG, 6GEI, 6GEK - PubMed Abstract:
Many interactions between proteins are mediated by intrinsically disordered regions (IDRs). Intrinsically disordered proteins (IDPs) do not adopt a stable three-dimensional structure in their unbound form, but they become more structured upon binding to their partners. In this communication, we study how a bound IDR adapts to mutations, preventing the formation of hydrogen bonds at the binding interface that needs a precise positioning of the interacting residues to be formed. We use as a model the YAP:TEAD interface, where one YAP (IDP) and two TEAD residues form hydrogen bonds via their side chain. Our study shows that the conformational flexibility of bound YAP and the reorganization of water molecules at the interface help to reduce the energetic constraints created by the loss of H-bonds at the interface. The residual flexibility/dynamic of bound IDRs and water might, therefore, be a key for the adaptation of IDPs to different interface landscapes and to mutations occurring at binding interfaces.
- Disease Area Oncology, Novartis Institutes for Biomedical Research, Basel, Switzerland.
Organizational Affiliation: