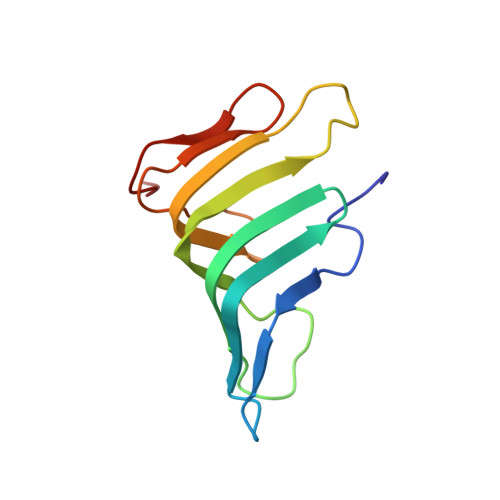
Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport inEscherichia coli.
Vetterli, S.U., Zerbe, K., Muller, M., Urfer, M., Mondal, M., Wang, S.Y., Moehle, K., Zerbe, O., Vitale, A., Pessi, G., Eberl, L., Wollscheid, B., Robinson, J.A.(2018) Sci Adv 4: eaau2634-eaau2634
- PubMed: 30443594
- DOI: https://doi.org/10.1126/sciadv.aau2634
- Primary Citation of Related Structures:
6GD5 - PubMed Abstract:
With the increasing resistance of many Gram-negative bacteria to existing classes of antibiotics, identifying new paradigms in antimicrobial discovery is an important research priority. Of special interest are the proteins required for the biogenesis of the asymmetric Gram-negative bacterial outer membrane (OM). Seven Lpt proteins (LptA to LptG) associate in most Gram-negative bacteria to form a macromolecular complex spanning the entire envelope, which transports lipopolysaccharide (LPS) molecules from their site of assembly at the inner membrane to the cell surface, powered by adenosine 5'-triphosphate hydrolysis in the cytoplasm. The periplasmic protein LptA comprises the protein bridge across the periplasm, which connects LptB 2 FGC at the inner membrane to LptD/E anchored in the OM. We show here that the naturally occurring, insect-derived antimicrobial peptide thanatin targets LptA and LptD in the network of periplasmic protein-protein interactions required to assemble the Lpt complex, leading to the inhibition of LPS transport and OM biogenesis in Escherichia coli .
- Chemistry Department, University of Zurich, Winterthurerstrasse 190, 8057 Zurich, Switzerland.
Organizational Affiliation: