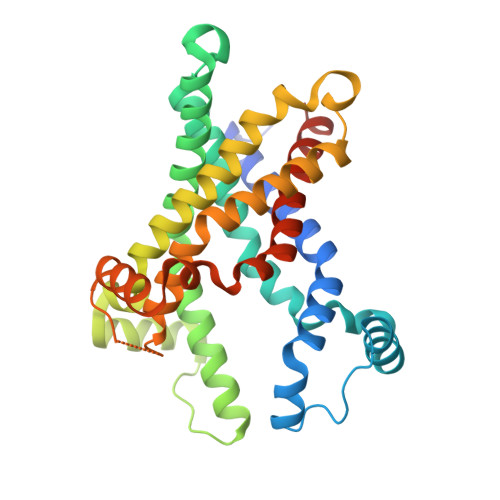
The Molecular Mechanism of Transport by the Mitochondrial ADP/ATP Carrier.
Ruprecht, J.J., King, M.S., Zogg, T., Aleksandrova, A.A., Pardon, E., Crichton, P.G., Steyaert, J., Kunji, E.R.S.(2019) Cell 176: 435-447.e15
- PubMed: 30611538
- DOI: https://doi.org/10.1016/j.cell.2018.11.025
- Primary Citation of Related Structures:
6GCI - PubMed Abstract:
Mitochondrial ADP/ATP carriers transport ADP into the mitochondrial matrix for ATP synthesis, and ATP out to fuel the cell, by cycling between cytoplasmic-open and matrix-open states. The structure of the cytoplasmic-open state is known, but it has proved difficult to understand the transport mechanism in the absence of a structure in the matrix-open state. Here, we describe the structure of the matrix-open state locked by bongkrekic acid bound in the ADP/ATP-binding site at the bottom of the central cavity. The cytoplasmic side of the carrier is closed by conserved hydrophobic residues, and a salt bridge network, braced by tyrosines. Glycine and small amino acid residues allow close-packing of helices on the matrix side. Uniquely, the carrier switches between states by rotation of its three domains about a fulcrum provided by the substrate-binding site. Because these features are highly conserved, this mechanism is likely to apply to the whole mitochondrial carrier family. VIDEO ABSTRACT.
- MRC Mitochondrial Biology Unit, University of Cambridge, Cambridge Biomedical Campus, Cambridge CB2 0XY, UK. Electronic address: jjr@mrc-mbu.cam.ac.uk.
Organizational Affiliation: