Structural basis for CD8+ T cells auto-reactivity in LCMV infection
Allerbring, E., Duru, A.D., Nygren, P.A., Sandalova, T., Achour, A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| H-2 class I histocompatibility antigen, D-B alpha chain | 276 | Mus musculus | Mutation(s): 0 Gene Names: H2-D1 | 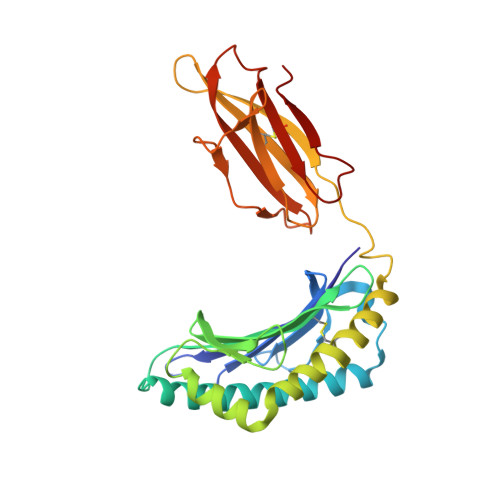 | |
UniProt | |||||
Find proteins for P01899 (Mus musculus) Explore P01899 Go to UniProtKB: P01899 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01899 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-2-microglobulin | 99 | Mus musculus | Mutation(s): 1 Gene Names: B2m | 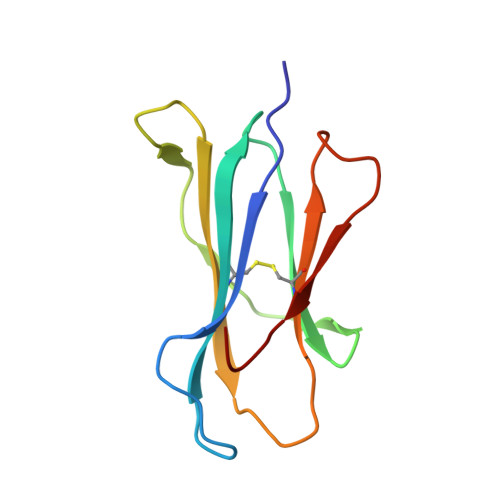 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01887 (Mus musculus) Explore P01887 Go to UniProtKB: P01887 | |||||
IMPC: MGI:88127 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01887 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Dopamine beta-hydroxylase | I [auth P], J [auth I], K [auth J], L [auth K] | 9 | Mus musculus | Mutation(s): 1 EC: 1.14.17.1 | 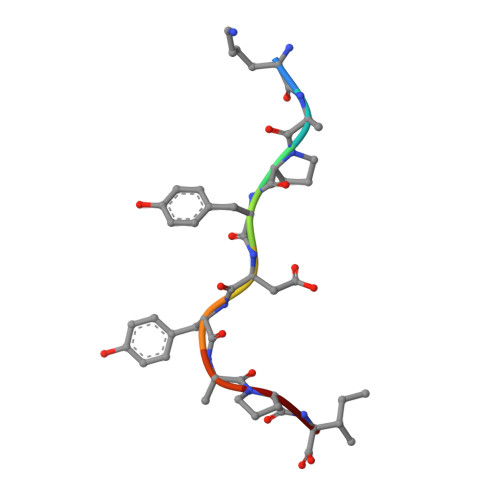 |
UniProt | |||||
Find proteins for Q64237 (Mus musculus) Explore Q64237 Go to UniProtKB: Q64237 | |||||
Entity Groups | |||||
| UniProt Group | Q64237 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 92.245 | α = 90 |
| b = 123.597 | β = 103.07 |
| c = 99.262 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |