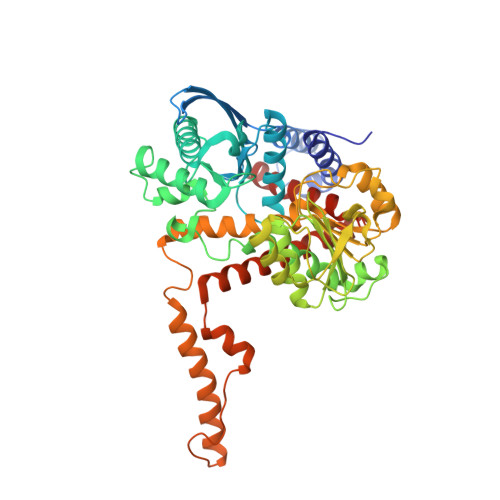
Crystal structure of glutamate dehydrogenase 2, a positively selected novel human enzyme involved in brain biology and cancer pathophysiology.
Dimovasili, C., Fadouloglou, V.E., Kefala, A., Providaki, M., Kotsifaki, D., Kanavouras, K., Sarrou, I., Plaitakis, A., Zaganas, I., Kokkinidis, M.(2021) J Neurochem
- PubMed: 33421122
- DOI: https://doi.org/10.1111/jnc.15296
- Primary Citation of Related Structures:
6G2U - PubMed Abstract:
Mammalian glutamate dehydrogenase (hGDH1 in human cells) interconverts glutamate to α-ketoglutarate and ammonia while reducing NAD(P) to NAD(P)H. During primate evolution, humans and great apes have acquired hGDH2, an isoenzyme that underwent rapid evolutionary adaptation concomitantly with brain expansion, thereby acquiring unique catalytic and regulatory properties that permitted its function under conditions inhibitory to its ancestor hGDH1. Although the 3D-structures of GDHs, including hGDH1, have been determined, attempts to determine the hGDH2 structure were until recently unsuccessful. Comparison of the hGDH1/hGDH2 structures would enable a detailed understanding of their evolutionary differences. This work aimed at the determination of the hGDH2 crystal structure and the analysis of its functional implications. Recombinant hGDH2 was produced in the Spodoptera frugiperda ovarian cell line Sf21, using the Baculovirus expression system. Purification was achieved via a two-step chromatography procedure. hGDH2 was crystallized, X-ray diffraction data were collected using synchrotron radiation and the structure was determined by molecular replacement. The hGDH2 structure is reported at a resolution of 2.9 Å. The enzyme adopts a novel semi-closed conformation, which is an intermediate between known open and closed GDH1 conformations, differing from both. The structure enabled us to dissect previously reported biochemical findings and to structurally interpret the effects of evolutionary amino acid substitutions, including Arg470His, on ADP affinity. In conclusion, our data provide insights into the structural basis of hGDH2 properties, the functional evolution of hGDH isoenzymes, and open new prospects for drug design, especially for cancer therapeutics.
- Neurology Laboratory, Medical School, University of Crete, Heraklion, Crete, Greece.
Organizational Affiliation: