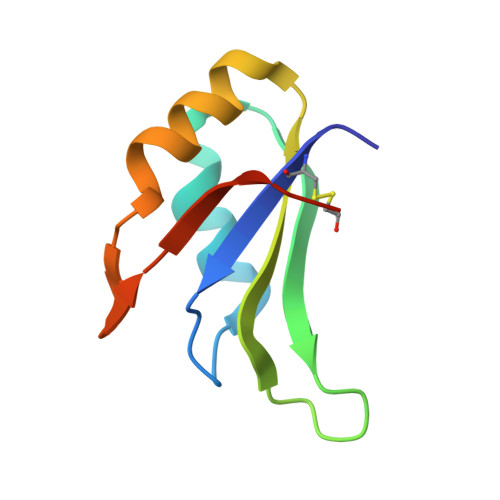
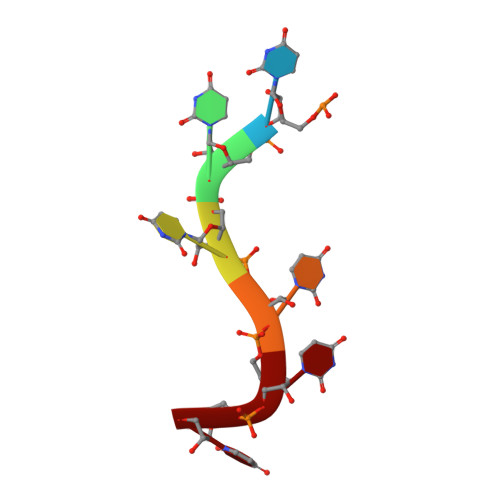
HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Pabis, M., Popowicz, G.M., Stehle, R., Fernandez-Ramos, D., Asami, S., Warner, L., Garcia-Maurino, S.M., Schlundt, A., Martinez-Chantar, M.L., Diaz-Moreno, I., Sattler, M.(2019) Nucleic Acids Res 47: 1011-1029
- PubMed: 30418581
- DOI: https://doi.org/10.1093/nar/gky1138
- Primary Citation of Related Structures:
6G2K, 6GD1, 6GD2, 6GD3 - PubMed Abstract:
HuR/ELAVL1 is an RNA-binding protein involved in differentiation and stress response that acts primarily by stabilizing messenger RNA (mRNA) targets. HuR comprises three RNA recognition motifs (RRMs) where the structure and RNA binding of RRM3 and of full-length HuR remain poorly understood. Here, we report crystal structures of RRM3 free and bound to cognate RNAs. Our structural, NMR and biochemical data show that RRM3 mediates canonical RNA interactions and reveal molecular details of a dimerization interface localized on the α-helical face of RRM3. NMR and SAXS analyses indicate that the three RRMs in full-length HuR are flexibly connected in the absence of RNA, while they adopt a more compact arrangement when bound to RNA. Based on these data and crystal structures of tandem RRM1,2-RNA and our RRM3-RNA complexes, we present a structural model of RNA recognition involving all three RRM domains of full-length HuR. Mutational analysis demonstrates that RRM3 dimerization and RNA binding is required for functional activity of full-length HuR in vitro and to regulate target mRNAs levels in human cells, thus providing a fine-tuning for HuR activity in vivo.
- Institute of Structural Biology, Helmholtz Zentrum München, Neuherberg, Germany.
Organizational Affiliation: