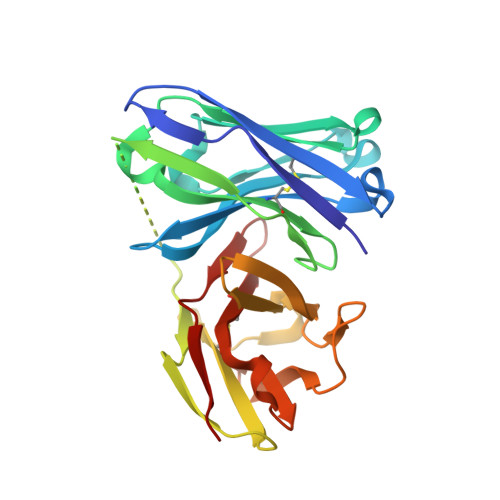
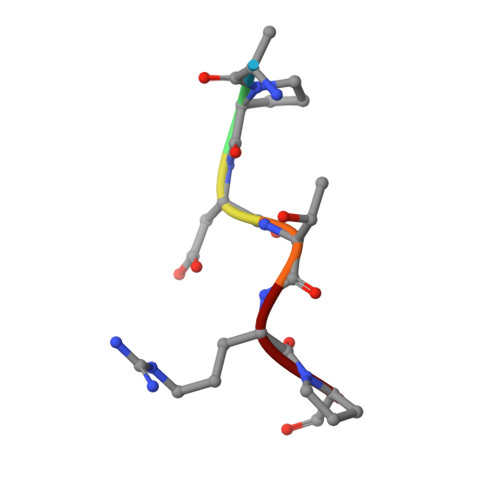
Water Sculpts the Distinctive Shapes and Dynamics of the Tumor-Associated Carbohydrate Tn Antigens: Implications for Their Molecular Recognition.
Bermejo, I.A., Usabiaga, I., Companon, I., Castro-Lopez, J., Insausti, A., Fernandez, J.A., Avenoza, A., Busto, J.H., Jimenez-Barbero, J., Asensio, J.L., Peregrina, J.M., Jimenez-Oses, G., Hurtado-Guerrero, R., Cocinero, E.J., Corzana, F.(2018) J Am Chem Soc 140: 9952-9960
- PubMed: 30004703
- DOI: https://doi.org/10.1021/jacs.8b04801
- Primary Citation of Related Structures:
6FZQ, 6FZR - PubMed Abstract:
The tumor-associated carbohydrate Tn antigens include two variants, αGalNAc- O-Thr and αGalNAc- O-Ser. In solution, they exhibit dissimilar shapes and dynamics and bind differently to the same protein receptor. Here, we demonstrate experimentally and theoretically that their conformational preferences in the gas phase are highly similar, revealing the essential role of water. We propose that water molecules prompt the rotation around the glycosidic linkage in the threonine derivative, shielding its hydrophobic methyl group and allowing an optimal solvation of the polar region of the antigen. The unusual arrangement of αGalNAc- O-Thr features a water molecule bound into a "pocket" between the sugar and the threonine. This mechanism is supported by trapping, for the first time, such localized water in the crystal structures of an antibody bound to two glycopeptides that comprise fluorinated Tn antigens in their structure. According to several reported X-ray structures, installing oxygenated amino acids in specific regions of the receptor capable of displacing the bridging water molecule to the bulk-solvent may facilitate the molecular recognition of the Tn antigen with threonine. Overall, our data also explain how water fine-tunes the 3D structure features of similar molecules, which in turn are behind their distinct biological activities.
- Departamento de Química, Centro de Investigación en Síntesis Química , Universidad de La Rioja , 26006 Logroño , Spain.
Organizational Affiliation: