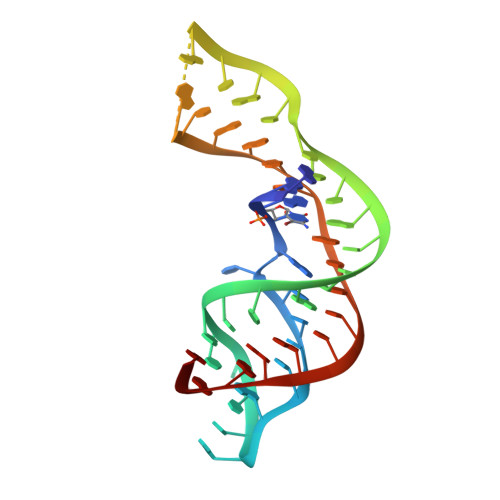
Structure and ligand binding of the SAM-V riboswitch.
Huang, L., Lilley, D.M.J.(2018) Nucleic Acids Res 46: 6869-6879
- PubMed: 29931337
- DOI: https://doi.org/10.1093/nar/gky520
- Primary Citation of Related Structures:
6FZ0 - PubMed Abstract:
SAM-V is one of the class of riboswitches that bind S-adenosylmethione, regulating gene expression by controlling translation. We have solved the crystal structure of the metY SAM-V riboswitch bound to its SAM ligand at 2.5 Å resolution. The RNA folds as an H-type pseudoknot, with a major-groove triple helix in which resides the SAM ligand binding site. The bound SAM adopts an elongated conformation aligned with the axis of the triple helix, and is held at either end by hydrogen bonding to the adenine and the amino acid moieties. The central sulfonium cation makes electrostatic interactions with an U:A.U base triple, so conferring specificity. We propose a model in which SAM binding leads to association of the triplex third strand that stabilizes a short helix and occludes the ribosome binding site. Thus the new structure explains both ligand specificity and the mechanism of genetic control.
- Cancer Research UK Nucleic Acid Structure Research Group, MSI/WTB Complex, The University of Dundee, Dow Street, Dundee DD1 5EH, UK.
Organizational Affiliation: