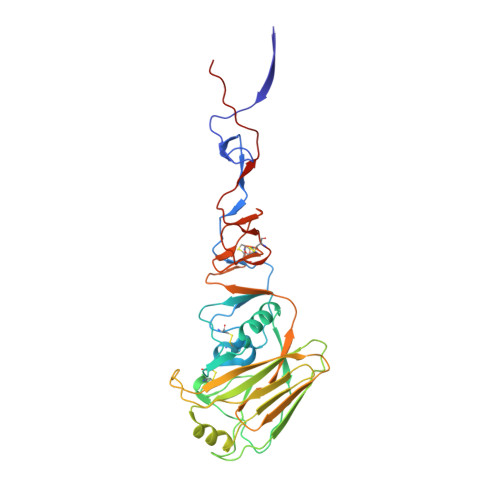
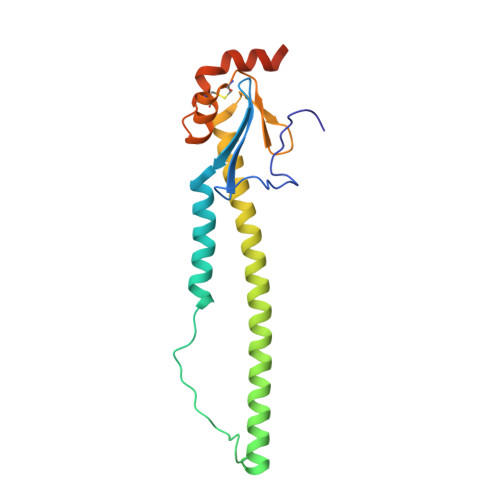
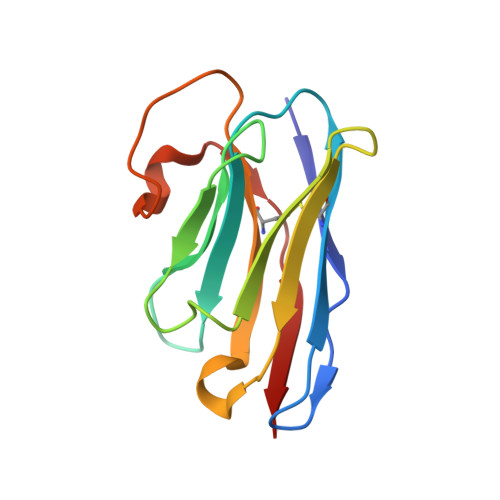
Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Laursen, N.S., Friesen, R.H.E., Zhu, X., Jongeneelen, M., Blokland, S., Vermond, J., van Eijgen, A., Tang, C., van Diepen, H., Obmolova, G., van der Neut Kolfschoten, M., Zuijdgeest, D., Straetemans, R., Hoffman, R.M.B., Nieusma, T., Pallesen, J., Turner, H.L., Bernard, S.M., Ward, A.B., Luo, J., Poon, L.L.M., Tretiakova, A.P., Wilson, J.M., Limberis, M.P., Vogels, R., Brandenburg, B., Kolkman, J.A., Wilson, I.A.(2018) Science 362: 598-602
- PubMed: 30385580
- DOI: https://doi.org/10.1126/science.aaq0620
- Primary Citation of Related Structures:
6CK8, 6CNV, 6CNW, 6FYS, 6FYT, 6FYU, 6FYW - PubMed Abstract:
Broadly neutralizing antibodies against highly variable pathogens have stimulated the design of vaccines and therapeutics. We report the use of diverse camelid single-domain antibodies to influenza virus hemagglutinin to generate multidomain antibodies with impressive breadth and potency. Multidomain antibody MD3606 protects mice against influenza A and B infection when administered intravenously or expressed locally from a recombinant adeno-associated virus vector. Crystal and single-particle electron microscopy structures of these antibodies with hemagglutinins from influenza A and B viruses reveal binding to highly conserved epitopes. Collectively, our findings demonstrate that multidomain antibodies targeting multiple epitopes exhibit enhanced virus cross-reactivity and potency. In combination with adeno-associated virus-mediated gene delivery, they may provide an effective strategy to prevent infection with influenza virus and other highly variable pathogens.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: