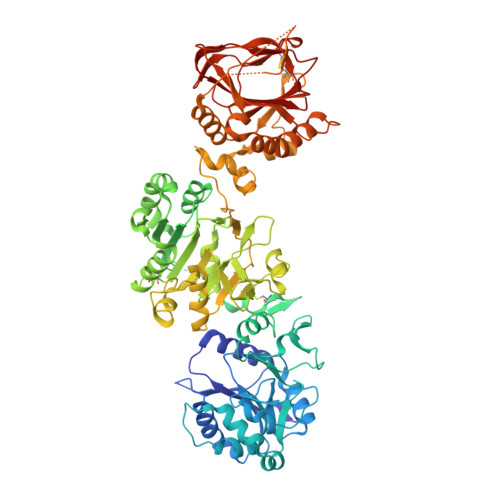
Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Scietti, L., Chiapparino, A., De Giorgi, F., Fumagalli, M., Khoriauli, L., Nergadze, S., Basu, S., Olieric, V., Cucca, L., Banushi, B., Profumo, A., Giulotto, E., Gissen, P., Forneris, F.(2018) Nat Commun 9: 3163-3163
- PubMed: 30089812
- DOI: https://doi.org/10.1038/s41467-018-05631-5
- Primary Citation of Related Structures:
6FXK, 6FXM, 6FXR, 6FXT, 6FXX, 6FXY - PubMed Abstract:
Lysyl hydroxylases catalyze hydroxylation of collagen lysines, and sustain essential roles in extracellular matrix (ECM) maturation and remodeling. Malfunctions in these enzymes cause severe connective tissue disorders. Human lysyl hydroxylase 3 (LH3/PLOD3) bears multiple enzymatic activities, as it catalyzes collagen lysine hydroxylation and also their subsequent glycosylation. Our understanding of LH3 functions is currently hampered by lack of molecular structure information. Here, we present high resolution crystal structures of full-length human LH3 in complex with cofactors and donor substrates. The elongated homodimeric LH3 architecture shows two distinct catalytic sites at the N- and C-terminal boundaries of each monomer, separated by an accessory domain. The glycosyltransferase domain displays distinguishing features compared to other known glycosyltransferases. Known disease-related mutations map in close proximity to the catalytic sites. Collectively, our results provide a structural framework characterizing the multiple functions of LH3, and the molecular mechanisms of collagen-related diseases involving human lysyl hydroxylases.
- The Armenise-Harvard Laboratory of Structural Biology, Department of Biology and Biotechnology, University of Pavia, Via Ferrata 9/A, 27100, Pavia, Italy.
Organizational Affiliation: