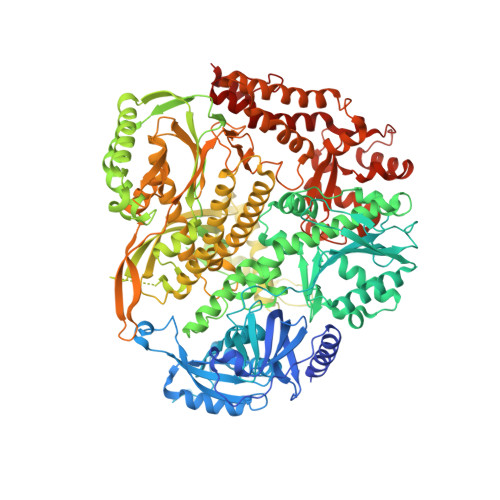
Structural consequence of the most frequently recurring cancer-associated substitution in DNA polymerase epsilon.
Parkash, V., Kulkarni, Y., Ter Beek, J., Shcherbakova, P.V., Kamerlin, S.C.L., Johansson, E.(2019) Nat Commun 10: 373-373
- PubMed: 30670696
- DOI: https://doi.org/10.1038/s41467-018-08114-9
- Primary Citation of Related Structures:
6FWK, 6G0A, 6I8A - PubMed Abstract:
The most frequently recurring cancer-associated DNA polymerase ε (Pol ε) mutation is a P286R substitution in the exonuclease domain. While originally proposed to increase genome instability by disrupting exonucleolytic proofreading, the P286R variant was later found to be significantly more pathogenic than Pol ε proofreading deficiency per se. The mechanisms underlying its stronger impact remained unclear. Here we report the crystal structure of the yeast orthologue, Pol ε-P301R, complexed with DNA and an incoming dNTP. Structural changes in the protein are confined to the exonuclease domain, with R301 pointing towards the exonuclease site. Molecular dynamics simulations suggest that R301 interferes with DNA binding to the exonuclease site, an outcome not observed with the exonuclease-inactive Pol ε-D290A,E292A variant lacking the catalytic residues. These results reveal a distinct mechanism of exonuclease inactivation by the P301R substitution and a likely basis for its dramatically higher mutagenic and tumorigenic effects.
- Department of Medical Biochemistry and Biophysics, Umeå University, Umeå, SE-90187, Sweden.
Organizational Affiliation: