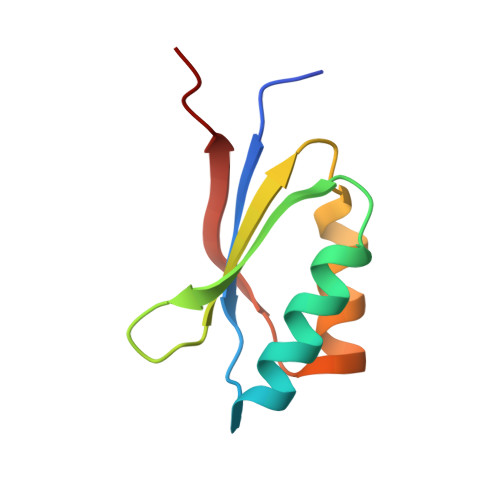
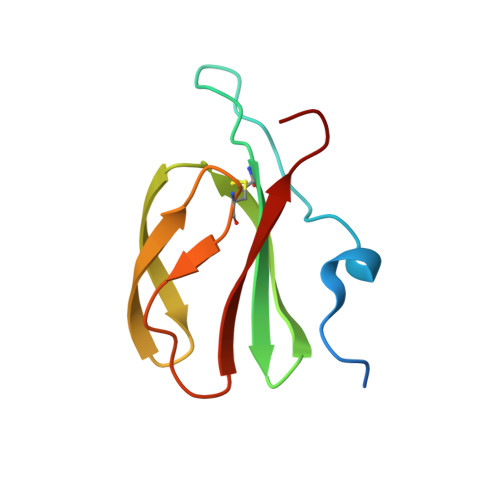
Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen.
De la Concepcion, J.C., Franceschetti, M., Maqbool, A., Saitoh, H., Terauchi, R., Kamoun, S., Banfield, M.J.(2018) Nat Plants 4: 576-585
- PubMed: 29988155
- DOI: https://doi.org/10.1038/s41477-018-0194-x
- Primary Citation of Related Structures:
6FU9, 6FUB, 6FUD, 6G10, 6G11 - PubMed Abstract:
Accelerated adaptive evolution is a hallmark of plant-pathogen interactions. Plant intracellular immune receptors (NLRs) often occur as allelic series with differential pathogen specificities. The determinants of this specificity remain largely unknown. Here, we unravelled the biophysical and structural basis of expanded specificity in the allelic rice NLR Pik, which responds to the effector AVR-Pik from the rice blast pathogen Magnaporthe oryzae. Rice plants expressing the Pikm allele resist infection by blast strains expressing any of three AVR-Pik effector variants, whereas those expressing Pikp only respond to one. Unlike Pikp, the integrated heavy metal-associated (HMA) domain of Pikm binds with high affinity to each of the three recognized effector variants, and variation at binding interfaces between effectors and Pikp-HMA or Pikm-HMA domains encodes specificity. By understanding how co-evolution has shaped the response profile of an allelic NLR, we highlight how natural selection drove the emergence of new receptor specificities. This work has implications for the engineering of NLRs with improved utility in agriculture.
- Department of Biological Chemistry, John Innes Centre, Norwich Research Park, Norwich, UK.
Organizational Affiliation: