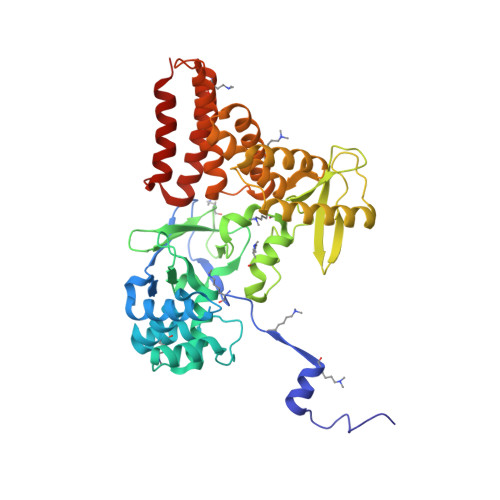
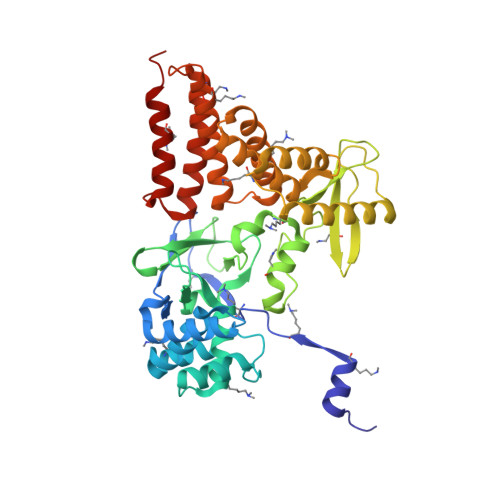
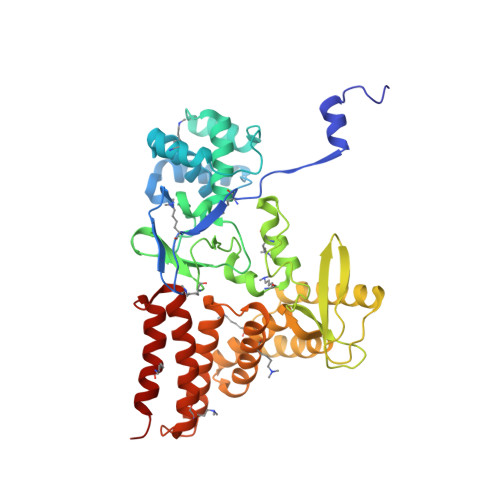
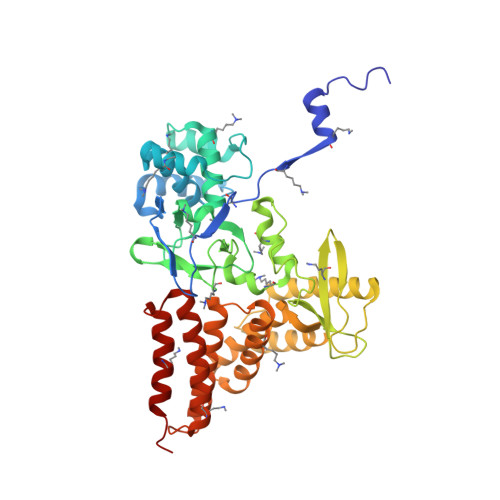
Structure of a Novel Dimeric SET Domain Methyltransferase that Regulates Cell Motility.
Pivovarova, Y., Liu, J., Lesigang, J., Koldyka, O., Rauschmeier, R., Hu, K., Dong, G.(2018) J Mol Biology 430: 4209-4229
- PubMed: 30148980
- DOI: https://doi.org/10.1016/j.jmb.2018.08.017
- Primary Citation of Related Structures:
6FND - PubMed Abstract:
Lysine methyltransferases (KMTs) were initially associated with transcriptional control through their methylation of histones and other nuclear proteins, but have since been found to regulate many other cellular activities. The apical complex lysine (K) methyltransferase (AKMT) of the human parasite Toxoplasma gondii was recently shown to play a critical role in regulating cellular motility. Here we report a 2.1-Å resolution crystal structure of the conserved and functional C-terminal portion (aa289-709) of T. gondii AKMT. AKMT dimerizes via a unique intermolecular interface mediated by the C-terminal tetratricopeptide repeat-like domain together with a specific zinc-binding motif that is absent from all other KMTs. Disruption of AKMT dimerization impaired both its enzyme activity and parasite egress from infected host cells in vivo. Structural comparisons reveal that AKMT is related to the KMTs in the SMYD family, with, however, a number of distinct structural features in addition to the unusual dimerization interface. These features are conserved among the apicomplexan parasites and their free-living relatives, but not found in any known KMTs in animals. AKMT therefore is the founding member of a new subclass of KMT that has important implications for the evolution of the apicomplexans.
- Max F. Perutz Laboratories, Medical University of Vienna, Vienna Biocenter (VBC), Dr. Bohr-Gasse 9, 1030 Vienna, Austria.
Organizational Affiliation: