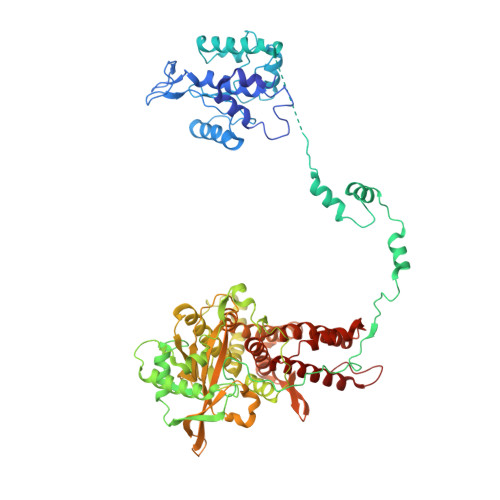
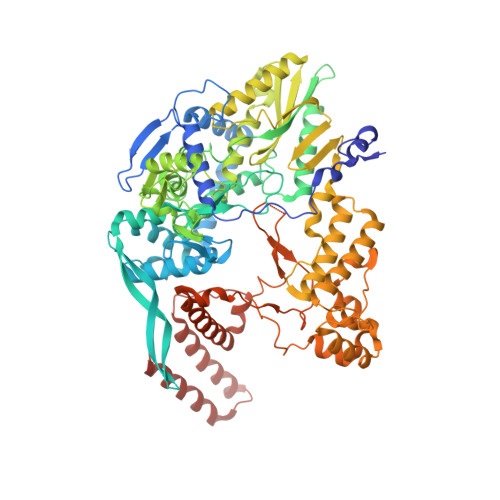
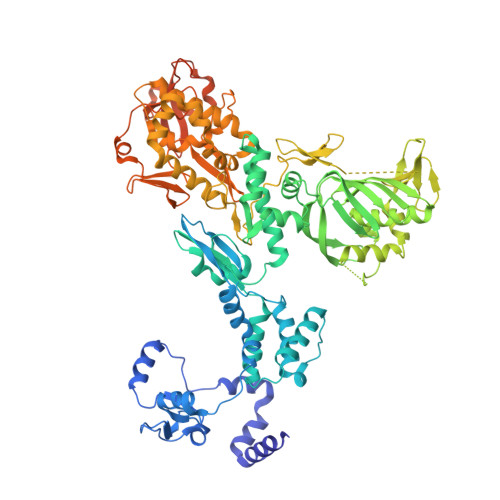
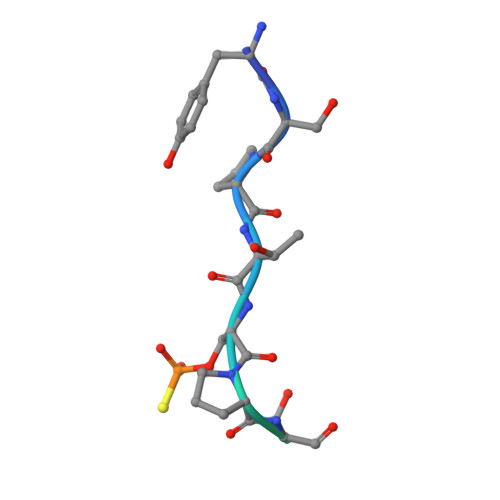
Towards New Anti-Influenza Therapeutics: Structure-Activity Relationships in the Interaction between Heterotrimeric Influenza Polymerase and Pol II C-terminal domain
Claron, M., Lukarska, M., Uhrig, U., Sehr, P., Drncova, P., Lewis, J.D., Will, D.W., Cusack, S.To be published.