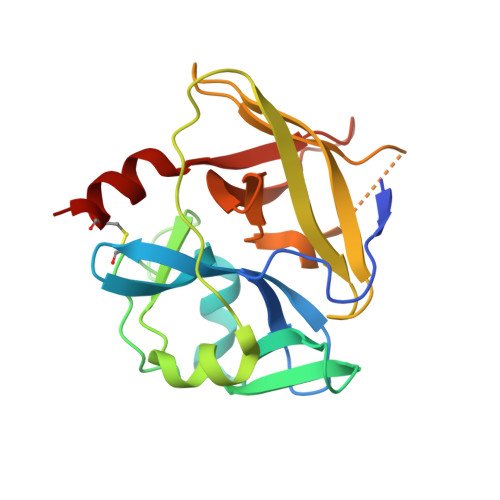
VPg Impact on Ryegrass Mottle Virus Serine-like 3C Protease Proteolysis and Structure.
Kalnins, G., Ludviga, R., Kalnciema, I., Resevica, G., Zeltina, V., Bogans, J., Tars, K., Zeltins, A., Balke, I.(2023) Int J Mol Sci 24
- PubMed: 36982419
- DOI: https://doi.org/10.3390/ijms24065347
- Primary Citation of Related Structures:
6FEZ, 6FF0, 7YZV - PubMed Abstract:
Sobemoviruses encode serine-like 3C proteases (Pro) that participate in the processing and maturation of other virus-encoded proteins. Its cis and trans activity is mediated by the naturally unfolded virus-genome-linked protein (VPg). Nuclear magnetic resonance studies show a Pro-VPg complex interaction and VPg tertiary structure; however, information regarding structural changes of the Pro-VPg complex during interaction is lacking. Here, we solved a full Pro-VPg 3D structure of ryegrass mottle virus (RGMoV) that demonstrates the structural changes in three different conformations due to VPg interaction with Pro. We identified a unique site of VPg interaction with Pro that was not observed in other sobemoviruses, and observed different conformations of the Pro β2 barrel. This is the first report of a full plant Pro crystal structure with its VPg cofactor. We also confirmed the existence of an unusual previously unmapped cleavage site for sobemovirus Pro in the transmembrane domain: E/A. We demonstrated that RGMoV Pro in cis activity is not regulated by VPg and that in trans , VPg can also mediate Pro in free form. Additionally, we observed Ca 2+ and Zn 2+ inhibitory effects on the Pro cleavage activity.
- Structural Biology Group, Latvian Biomedical Research and Study Centre, Ratsupites Street 1, k-1, LV-1067 Riga, Latvia.
Organizational Affiliation: