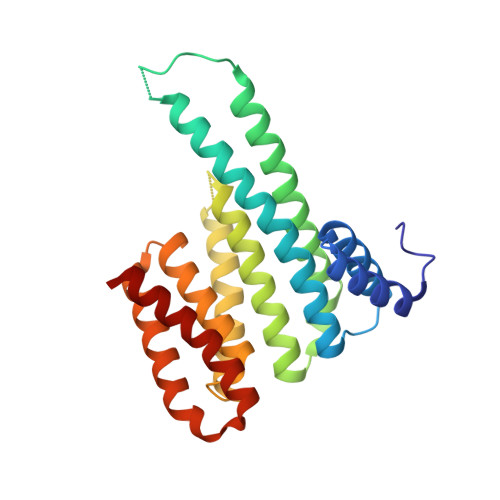
Characterization of Coding/Noncoding Variants forSHROOM3in Patients with CKD.
Prokop, J.W., Yeo, N.C., Ottmann, C., Chhetri, S.B., Florus, K.L., Ross, E.J., Sosonkina, N., Link, B.A., Freedman, B.I., Coppola, C.J., McDermott-Roe, C., Leysen, S., Milroy, L.G., Meijer, F.A., Geurts, A.M., Rauscher, F.J., Ramaker, R., Flister, M.J., Jacob, H.J., Mendenhall, E.M., Lazar, J.(2018) J Am Soc Nephrol 29: 1525-1535
- PubMed: 29476007
- DOI: https://doi.org/10.1681/ASN.2017080856
- Primary Citation of Related Structures:
6FBB, 6FCP - PubMed Abstract:
Background Interpreting genetic variants is one of the greatest challenges impeding analysis of rapidly increasing volumes of genomic data from patients. For example, SHROOM3 is an associated risk gene for CKD, yet causative mechanism(s) of SHROOM3 allele(s) are unknown. Methods We used our analytic pipeline that integrates genetic, computational, biochemical, CRISPR/Cas9 editing, molecular, and physiologic data to characterize coding and noncoding variants to study the human SHROOM3 risk locus for CKD. Results We identified a novel SHROOM3 transcriptional start site, which results in a shorter isoform lacking the PDZ domain and is regulated by a common noncoding sequence variant associated with CKD (rs17319721, allele frequency: 0.35). This variant disrupted allele binding to the transcription factor TCF7L2 in podocyte cell nuclear extracts and altered transcription levels of SHROOM3 in cultured cells, potentially through the loss of repressive looping between rs17319721 and the novel start site. Although common variant mechanisms are of high utility, sequencing is beginning to identify rare variants involved in disease; therefore, we used our biophysical tools to analyze an average of 112,849 individual human genome sequences for rare SHROOM3 missense variants, revealing 35 high-effect variants. The high-effect alleles include a coding variant (P1244L) previously associated with CKD ( P =0.01, odds ratio=7.95; 95% CI, 1.53 to 41.46) that we find to be present in East Asian individuals at an allele frequency of 0.0027. We determined that P1244L attenuates the interaction of SHROOM3 with 14-3-3, suggesting alterations to the Hippo pathway, a known mediator of CKD. Conclusions These data demonstrate multiple new SHROOM3 -dependent genetic/molecular mechanisms that likely affect CKD.
- HudsonAlpha Institute for Biotechnology, Huntsville, Alabama; jprokop54@gmail.com jlazar@hudsonalpha.org.
Organizational Affiliation: