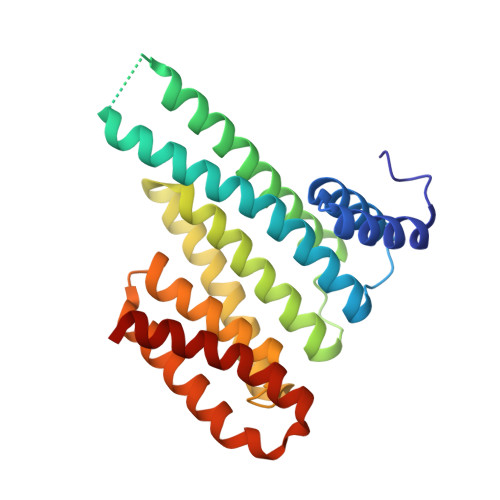
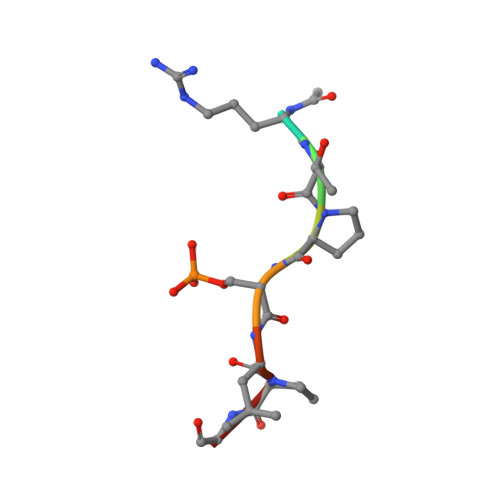
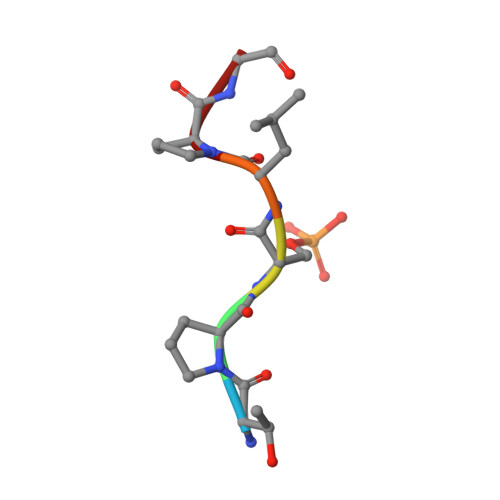
Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
Andrei, S.A., Meijer, F.A., Neves, J.F., Brunsveld, L., Landrieu, I., Ottmann, C., Milroy, L.G.(2018) ACS Chem Neurosci 9: 2639-2654
- PubMed: 29722962
- DOI: https://doi.org/10.1021/acschemneuro.8b00118
- Primary Citation of Related Structures:
6FAU, 6FAV, 6FAW, 6FBW, 6FBY, 6FI4, 6FI5 - PubMed Abstract:
Current molecular hypotheses have not yet delivered marketable treatments for Alzheimer's disease (AD), arguably due to a lack of understanding of AD biology and an overreliance on conventional drug modalities. Protein-protein interactions (PPIs) are emerging drug targets, which show promise for the treatment of, e.g., cancer, but are still underexploited for treating neurodegenerative diseases. 14-3-3 binding to phosphorylated Tau is a promising PPI drug target based on its reported destabilizing effect on microtubules, leading to enhanced neurofibrillary tangle formation as a potential cause of AD-related neurodegeneration. Inhibition of 14-3-3/Tau may therefore be neuroprotective. Previously, we reported the structure-guided development of modified peptide inhibitors of 14-3-3/Tau. Here, we report further efforts to optimize the binding mode and activity of our modified Tau peptides through a combination of chemical synthesis, biochemical assays, and X-ray crystallography. Most notably, we were able to characterize two different high-affinity binding modes, both of which inhibited 14-3-3-binding to full-length PKA-phosphorylated Tau protein in vitro as measured by NMR spectroscopy. Our findings, besides producing useful tool inhibitor compounds for studying 14-3-3/Tau, have enhanced our understanding of the molecular parameters for inhibiting 14-3-3/Tau, which are important milestones toward the establishment of our 14-3-3 PPI hypothesis.
- Laboratory of Chemical Biology, Department of Biomedical Engineering and Institute for Complex Molecular Systems , Technische Universiteit Eindhoven , Den Dolech 2 , 5612 AZ Eindhoven , The Netherlands.
Organizational Affiliation: