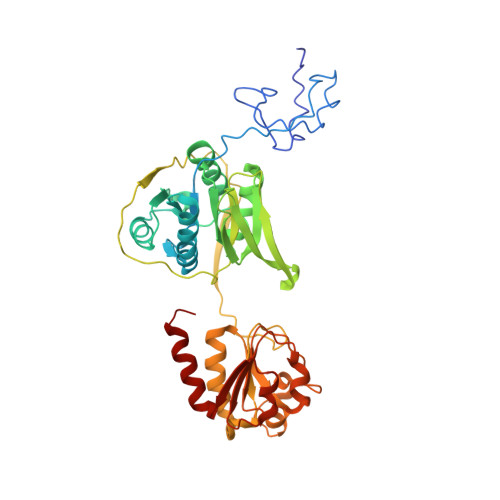
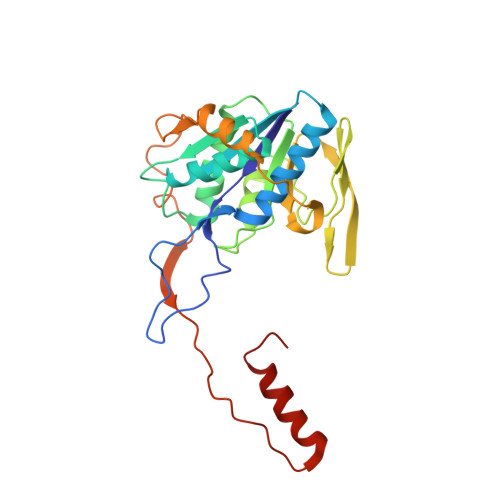
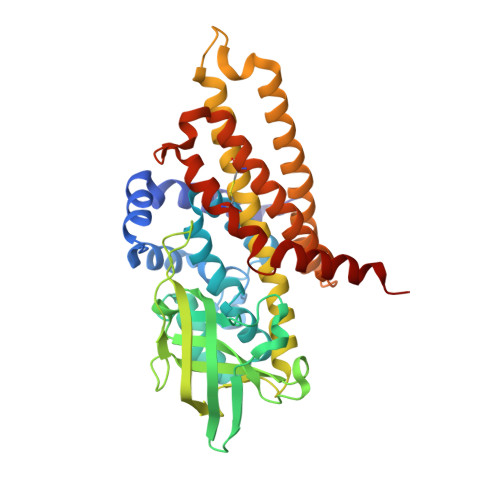
Molecular basis of the flavin-based electron-bifurcating caffeyl-CoA reductase reaction.
Demmer, J.K., Bertsch, J., Oppinger, C., Wohlers, H., Kayastha, K., Demmer, U., Ermler, U., Muller, V.(2018) FEBS Lett 592: 332-342
- PubMed: 29325219
- DOI: https://doi.org/10.1002/1873-3468.12971
- Primary Citation of Related Structures:
6FAH - PubMed Abstract:
Flavin-based electron bifurcation (FBEB) is a recently discovered mode of energy coupling in anaerobic microorganisms. The electron-bifurcating caffeyl-CoA reductase (CarCDE) catalyzes the reduction of caffeyl-CoA and ferredoxin by oxidizing NADH. The 3.5 Å structure of the heterododecameric Car(CDE) 4 complex of Acetobacterium woodii, presented here, reveals compared to other electron-transferring flavoprotein/acyl dehydrogenase family members an additional ferredoxin-like domain with two [4Fe-4S] clusters N-terminally fused to CarE. It might serve, in vivo, as specific adaptor for the physiological electron acceptor. Kinetic analysis of a CarCDE(∆Fd) complex indicates the bypassing of the ferredoxin-like domain by artificial electron acceptors. Site-directed mutagenesis studies substantiated the crucial role of the C-terminal arm of CarD and of ArgE203, hydrogen-bonded to the bifurcating FAD, for FBEB.
- Max-Planck-Institut für Biophysik, Frankfurt am Main, Germany.
Organizational Affiliation: