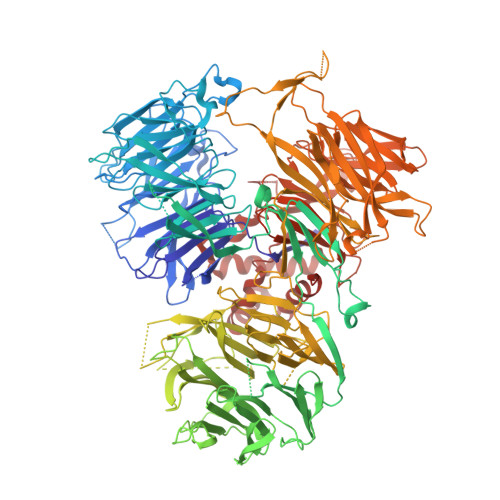
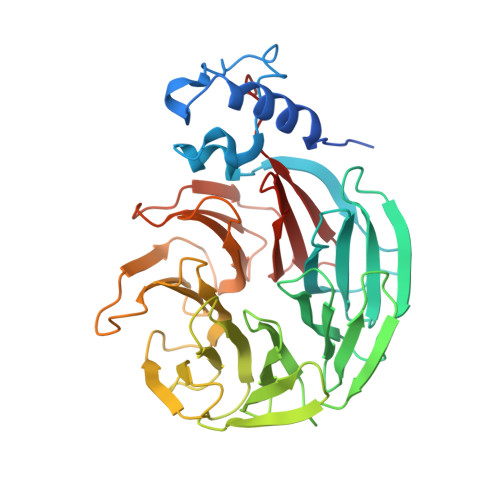
Structural insights into the assembly and polyA signal recognition mechanism of the human CPSF complex.
Clerici, M., Faini, M., Aebersold, R., Jinek, M.(2017) Elife 6
- PubMed: 29274231
- DOI: https://doi.org/10.7554/eLife.33111
- Primary Citation of Related Structures:
6F9N - PubMed Abstract:
3' polyadenylation is a key step in eukaryotic mRNA biogenesis. In mammalian cells, this process is dependent on the recognition of the hexanucleotide AAUAAA motif in the pre-mRNA polyadenylation signal by the cleavage and polyadenylation specificity factor (CPSF) complex. A core CPSF complex comprising CPSF160, WDR33, CPSF30 and Fip1 is sufficient for AAUAAA motif recognition, yet the molecular interactions underpinning its assembly and mechanism of PAS recognition are not understood. Based on cross-linking-coupled mass spectrometry, crystal structure of the CPSF160-WDR33 subcomplex and biochemical assays, we define the molecular architecture of the core human CPSF complex, identifying specific domains involved in inter-subunit interactions. In addition to zinc finger domains in CPSF30, we identify using quantitative RNA-binding assays an N-terminal lysine/arginine-rich motif in WDR33 as a critical determinant of specific AAUAAA motif recognition. Together, these results shed light on the function of CPSF in mediating PAS-dependent RNA cleavage and polyadenylation.
- Department of Biochemistry, University of Zurich, Zurich, Switzerland.
Organizational Affiliation: