A Mechanism for the Activation of the Influenza Virus Transcriptase.
Serna Martin, I., Hengrung, N., Renner, M., Sharps, J., Martinez-Alonso, M., Masiulis, S., Grimes, J.M., Fodor, E.(2018) Mol Cell 70: 1101-1110.e4
- PubMed: 29910112
- DOI: https://doi.org/10.1016/j.molcel.2018.05.011
- Primary Citation of Related Structures:
6F5O, 6F5P - PubMed Abstract:
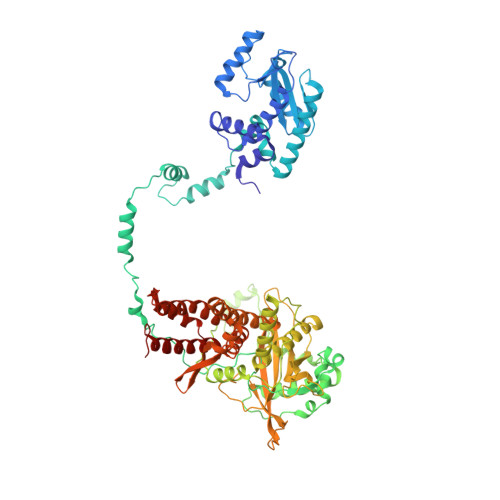
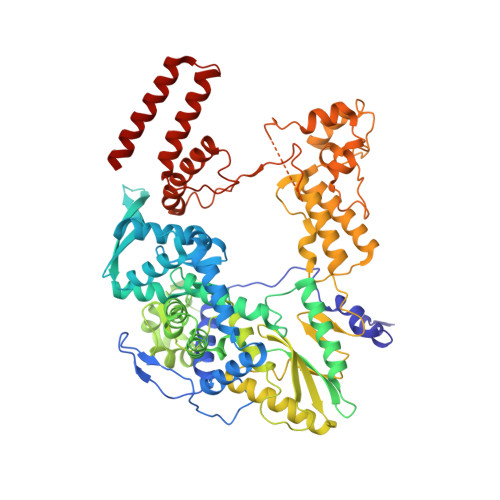
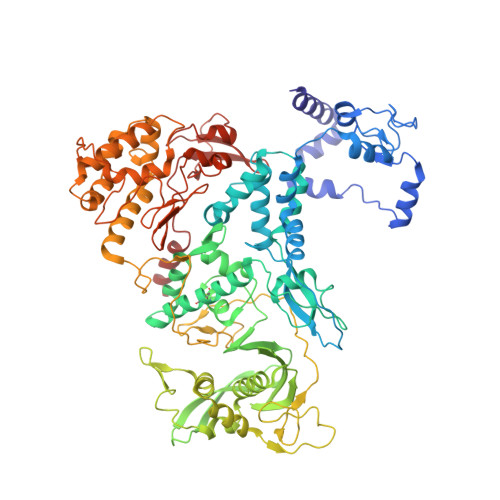
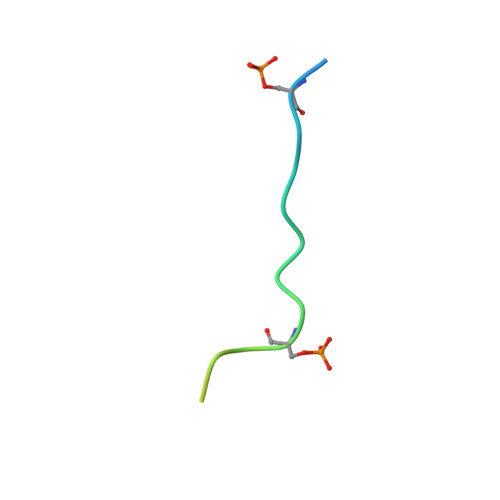
Influenza virus RNA polymerase (FluPol), a heterotrimer composed of PB1, PB2, and PA subunits (P3 in influenza C), performs both transcription and replication of the viral RNA genome. For transcription, FluPol interacts with the C-terminal domain (CTD) of RNA polymerase II (Pol II), which enables FluPol to snatch capped RNA primers from nascent host RNAs. Here, we describe the co-crystal structure of influenza C virus polymerase (FluPol C ) bound to a Ser5-phosphorylated CTD (pS 5 -CTD) peptide. The position of the CTD-binding site at the interface of PB1, P3, and the flexible PB2 C-terminal domains suggests that CTD binding stabilizes the transcription-competent conformation of FluPol. In agreement, both cap snatching and capped primer-dependent transcription initiation by FluPol C are enhanced in the presence of pS 5 -CTD. Mutations of amino acids in the CTD-binding site reduce viral mRNA synthesis. We propose a model for the activation of the influenza virus transcriptase through its association with pS 5 -CTD of Pol II.
- Sir William Dunn School of Pathology, University of Oxford, South Parks Road, Oxford OX1 3RE, UK.
Organizational Affiliation: