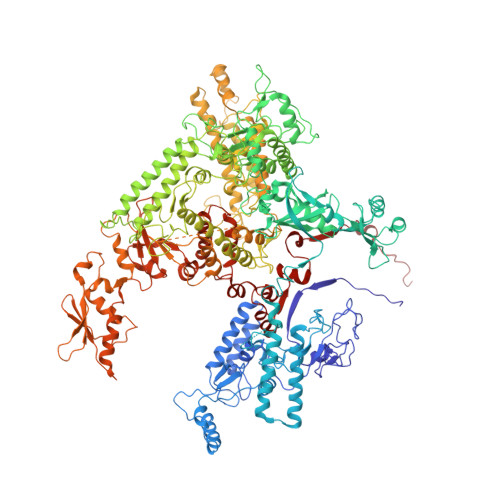
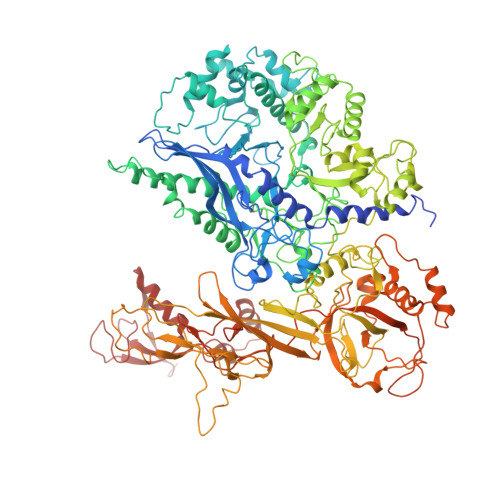
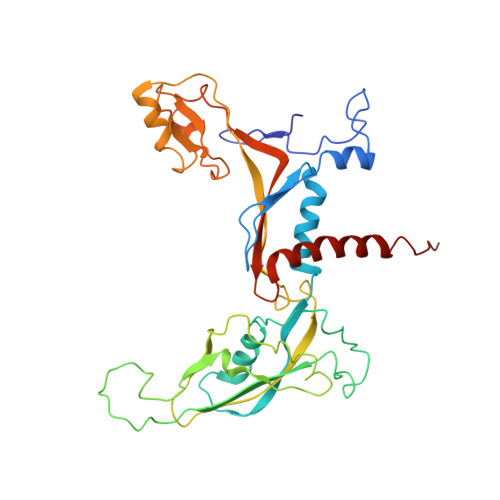
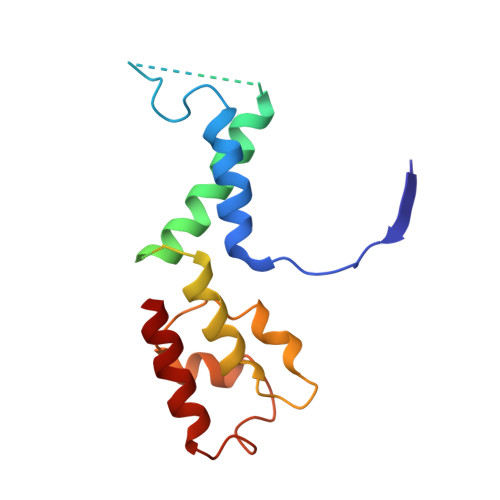
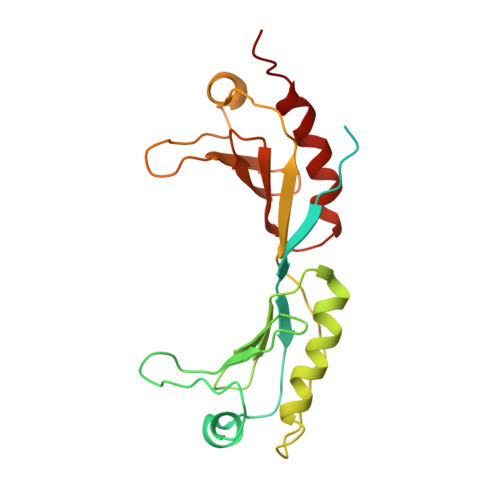
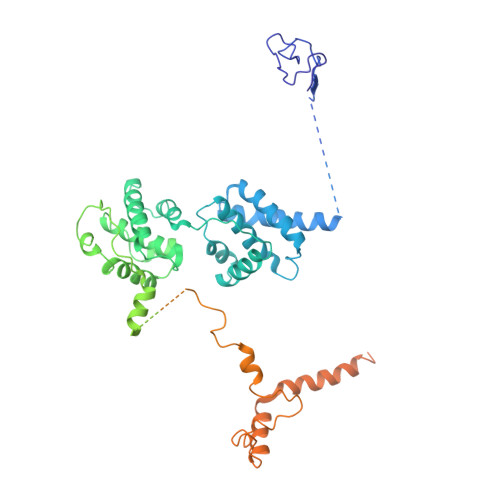
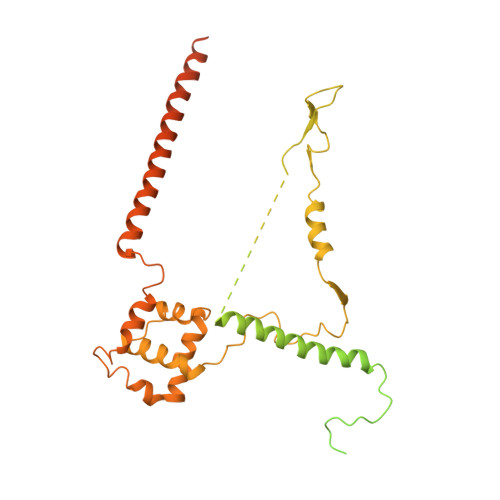
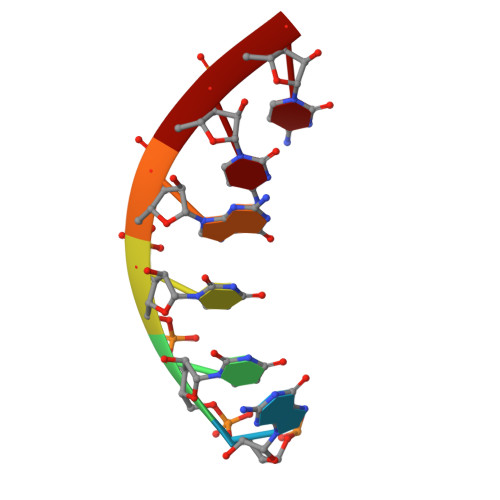
Molecular mechanism of promoter opening by RNA polymerase III.
Vorlander, M.K., Khatter, H., Wetzel, R., Hagen, W.J.H., Muller, C.W.(2018) Nature 553: 295-300
- PubMed: 29345638
- DOI: https://doi.org/10.1038/nature25440
- Primary Citation of Related Structures:
6F40, 6F41, 6F42, 6F44 - PubMed Abstract:
RNA polymerase III (Pol III) and transcription factor IIIB (TFIIIB) assemble together on different promoter types to initiate the transcription of small, structured RNAs. Here we present structures of Pol III preinitiation complexes, comprising the 17-subunit Pol III and the heterotrimeric transcription factor TFIIIB, bound to a natural promoter in different functional states. Electron cryo-microscopy reconstructions, varying from 3.7 Å to 5.5 Å resolution, include two early intermediates in which the DNA duplex is closed, an open DNA complex, and an initially transcribing complex with RNA in the active site. Our structures reveal an extremely tight, multivalent interaction between TFIIIB and promoter DNA, and explain how TFIIIB recruits Pol III. Together, TFIIIB and Pol III subunit C37 activate the intrinsic transcription factor-like activity of the Pol III-specific heterotrimer to initiate the melting of double-stranded DNA, in a mechanism similar to that of the Pol II system.
- European Molecular Biology Laboratory (EMBL), Structural and Computational Biology Unit, Meyerhofstrasse 1, 69117 Heidelberg, Germany.
Organizational Affiliation: