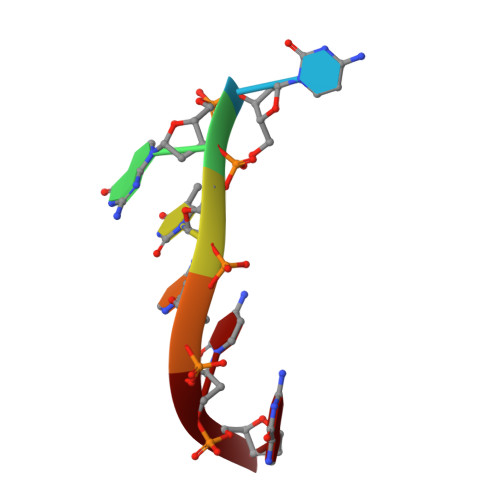
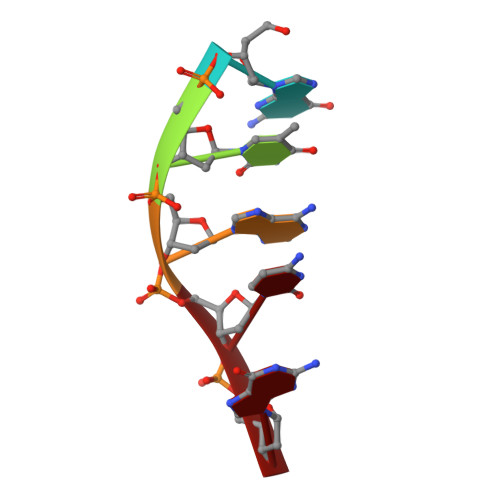
Induction of a Four-Way Junction Structure in the DNA Palindromic Hexanucleotide 5'-d(CGTACG)-3' by a Mononuclear Platinum Complex.
van Rixel, V.H.S., Busemann, A., Wissingh, M.F., Hopkins, S.L., Siewert, B., van de Griend, C., Siegler, M.A., Marzo, T., Papi, F., Ferraroni, M., Gratteri, P., Bazzicalupi, C., Messori, L., Bonnet, S.(2019) Angew Chem Int Ed Engl 58: 9378-9382
- PubMed: 31046177
- DOI: https://doi.org/10.1002/anie.201814532
- Primary Citation of Related Structures:
6F3C - PubMed Abstract:
Four-way junctions (4WJs) are supramolecular DNA assemblies comprising four interacting DNA strands that in biology are involved in DNA-damage repair. In this study, a new mononuclear platinum(II) complex 1 was prepared that is capable of driving the crystallization of the DNA oligomer 5'-d(CGTACG)-3' specifically into a 4WJ-like motif. In the crystal structure of the 1-CGTACG adduct, the distorted-square-planar platinum complex binds to the core of the 4WJ-like motif through π-π stacking and hydrogen bonding, without forming any platinum-nitrogen coordination bonds. Our observations suggest that the specific molecular properties of the metal complex are crucially responsible for triggering the selective assembly of this peculiar DNA superstructure.
- Leiden Institute of Chemistry, Leiden University, Einsteinweg 55, PO Box 9502, 2333CC, Leiden, The Netherlands.
Organizational Affiliation: