Multimerization of HIV-1 integrase hinges on conserved SH3-docking platforms
Galilee, M., Alian, A.(2018) bioRxiv
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2018) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Integrase | 152 | Human immunodeficiency virus 1 | Mutation(s): 0 Gene Names: pol | 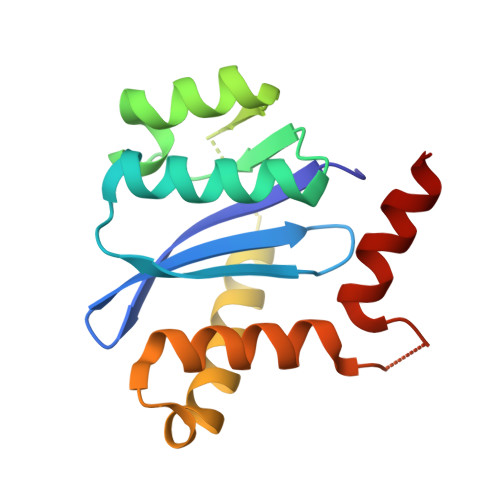 | |
UniProt | |||||
Find proteins for P04585 (Human immunodeficiency virus type 1 group M subtype B (isolate HXB2)) Explore P04585 Go to UniProtKB: P04585 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04585 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Inhibitor Peptide | 15 | synthetic construct | Mutation(s): 0 | 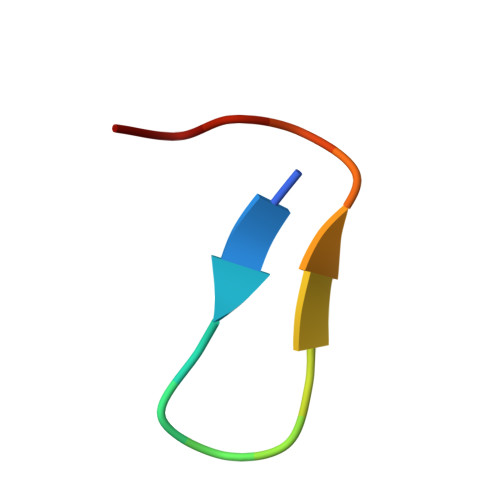 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 96.519 | α = 90 |
| b = 96.519 | β = 90 |
| c = 48.489 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| MOSFLM | data reduction |
| Aimless | data scaling |
| REFMAC | phasing |