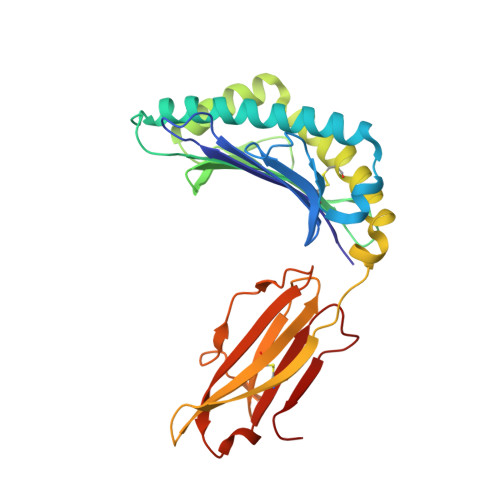
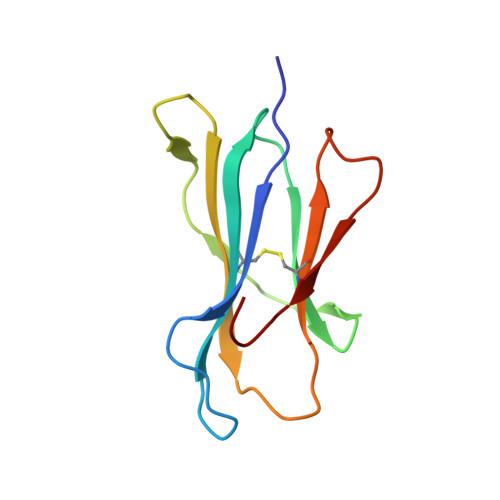
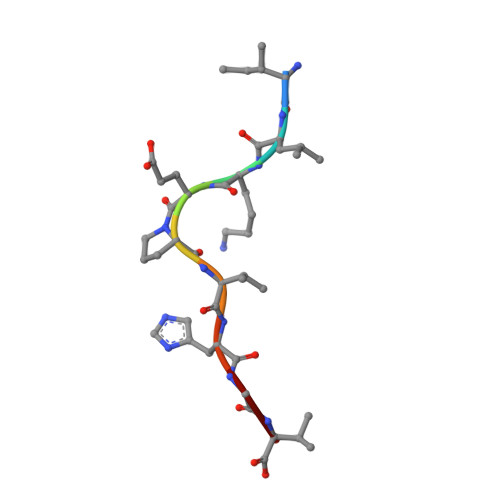
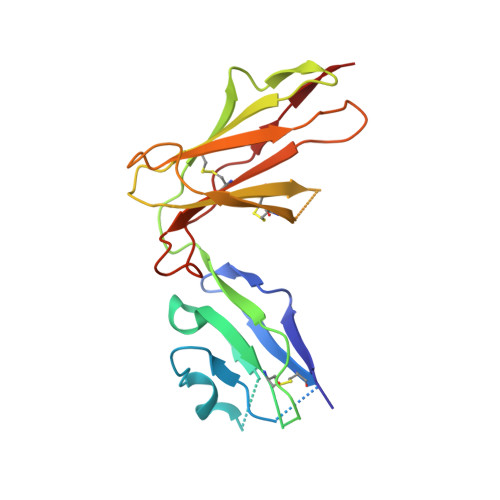
Application of the immunoregulatory receptor LILRB1 as a crystallisation chaperone for human class I MHC complexes.
Mohammed, F., Stones, D.H., Willcox, B.E.(2019) J Immunol Methods 464: 47-56
- PubMed: 30365927
- DOI: https://doi.org/10.1016/j.jim.2018.10.011
- Primary Citation of Related Structures:
6EWA, 6EWC, 6EWO - PubMed Abstract:
X-ray crystallographic studies of class I peptide-MHC molecules (pMHC) continue to provide important insights into immune recognition, however their success depends on generation of diffraction-quality crystals, which remains a significant challenge. While protein engineering techniques such as surface-entropy reduction and lysine methylation have proven utility in facilitating and/or improving protein crystallisation, they risk affecting the conformation and biochemistry of the class I MHC antigen binding groove. An attractive alternative is the use of noncovalent crystallisation chaperones, however these have not been developed for pMHC. Here we describe a method for promoting class I pMHC crystallisation, by exploiting its natural ligand interaction with the immunoregulatory receptor LILRB1 as a novel crystallisation chaperone. First, focussing on a model HIV-1-derived HLA-A2-restricted peptide, we determined a 2.4 Å HLA-A2/LILRB1 structure, which validated that co-crystallisation with LILRB1 does not alter conformation of the antigenic peptide. We then demonstrated that addition of LILRB1 enhanced the crystallisation of multiple peptide-HLA-A2 complexes, and identified a generic condition for initial co-crystallisation. LILRB1 chaperone-based crystallisation enabled structure determination for HLA-A2 complexes previously intransigent to crystallisation, including both conventional and post-translationally-modified peptides, of diverse lengths. Since both the LILRB1 recognition interface on the HLA-A2 α3 domain molecule and HLA-A2-mediated crystal contacts are predominantly conserved across class I MHC molecules, the approach we outline could prove applicable to a diverse range of class I pMHC. LILRB1 chaperone-mediated crystallisation should expedite molecular insights into the immunobiology of diverse immune-related diseases and immunotherapeutic strategies, particularly involving class I pMHC complexes that are challenging to crystallise.
- Cancer Immunology and Immunotherapy Centre, Institute of Immunology and Immunotherapy, University of Birmingham, Edgbaston, Birmingham B15 2TT, UK.
Organizational Affiliation: