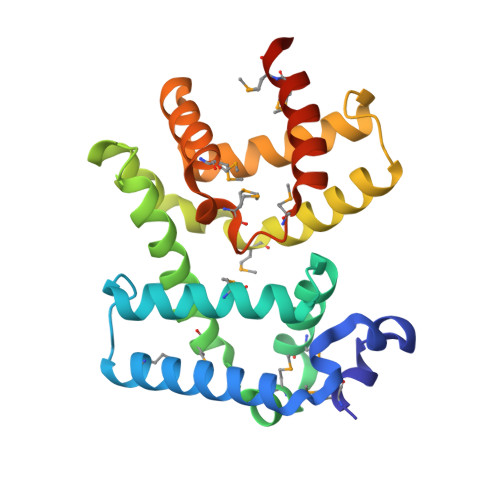
Molecular insights into antibiotic resistance - how a binding protein traps albicidin.
Rostock, L., Driller, R., Gratz, S., Kerwat, D., von Eckardstein, L., Petras, D., Kunert, M., Alings, C., Schmitt, F.J., Friedrich, T., Wahl, M.C., Loll, B., Mainz, A., Sussmuth, R.D.(2018) Nat Commun 9: 3095-3095
- PubMed: 30082794
- DOI: https://doi.org/10.1038/s41467-018-05551-4
- Primary Citation of Related Structures:
6ET8 - PubMed Abstract:
The worldwide emergence of antibiotic resistance poses a serious threat to human health. A molecular understanding of resistance strategies employed by bacteria is obligatory to generate less-susceptible antibiotics. Albicidin is a highly potent antibacterial compound synthesized by the plant-pathogenic bacterium Xanthomonas albilineans. The drug-binding protein AlbA confers albicidin resistance to Klebsiella oxytoca. Here we show that AlbA binds albicidin with low nanomolar affinity resulting in full inhibition of its antibacterial activity. We report on the crystal structure of the drug-binding domain of AlbA (AlbAS) in complex with albicidin. Both α-helical repeat domains of AlbAS are required to cooperatively clamp albicidin, which is unusual for drug-binding proteins of the MerR family. Structure-guided NMR binding studies employing synthetic albicidin derivatives give valuable information about ligand promiscuity of AlbAS. Our findings thus expand the general understanding of antibiotic resistance mechanisms and support current drug-design efforts directed at more effective albicidin analogs.
- Institut für Chemie, Biologische Chemie, Technische Universität Berlin, Straße des 17. Juni 124, 10623, Berlin, Germany.
Organizational Affiliation: