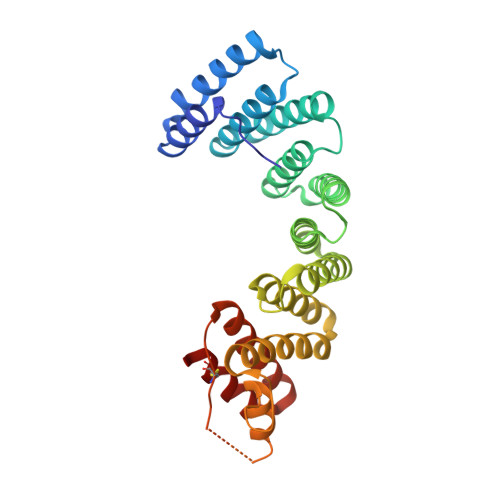
Insights into Kinesin-1 Activation from the Crystal Structure of KLC2 Bound to JIP3.
Cockburn, J.J.B., Hesketh, S.J., Mulhair, P., Thomsen, M., O'Connell, M.J., Way, M.(2018) Structure 26: 1486-1498.e6
- PubMed: 30197037
- DOI: https://doi.org/10.1016/j.str.2018.07.011
- Primary Citation of Related Structures:
6EJN, 6F9I - PubMed Abstract:
Kinesin-1 transports numerous cellular cargoes along microtubules. The kinesin-1 light chain (KLC) mediates cargo binding and regulates kinesin-1 motility. To investigate the molecular basis for kinesin-1 recruitment and activation by cargoes, we solved the crystal structure of the KLC2 tetratricopeptide repeat (TPR) domain bound to the cargo JIP3. This, combined with biophysical and molecular evolutionary analyses, reveals a kinesin-1 cargo binding site, located on KLC TPR1, which is conserved in homologs from sponges to humans. In the complex, JIP3 crosslinks two KLC2 TPR domains via their TPR1s. We show that TPR1 forms a dimer interface that mimics JIP3 binding in all crystal structures of the unbound KLC TPR domain. We propose that cargo-induced dimerization of the KLC TPR domains via TPR1 is a general mechanism for activating kinesin-1. We relate this to activation by tryptophan-acidic cargoes, explaining how different cargoes activate kinesin-1 through related molecular mechanisms.
- Astbury Centre for Structural Molecular Biology, University of Leeds, Leeds LS2 9JT, UK. Electronic address: j.j.b.cockburn@leeds.ac.uk.
Organizational Affiliation: