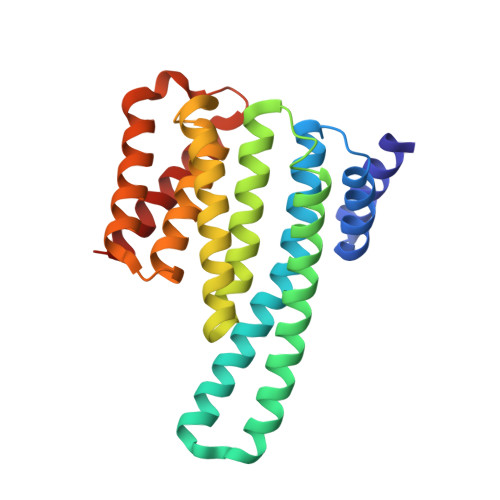
p38-MK2 signaling axis regulates RNA metabolism after UV-light-induced DNA damage.
Borisova, M.E., Voigt, A., Tollenaere, M.A.X., Sahu, S.K., Juretschke, T., Kreim, N., Mailand, N., Choudhary, C., Bekker-Jensen, S., Akutsu, M., Wagner, S.A., Beli, P.(2018) Nat Commun 9: 1017-1017
- PubMed: 29523821
- DOI: https://doi.org/10.1038/s41467-018-03417-3
- Primary Citation of Related Structures:
6EIH - PubMed Abstract:
Ultraviolet (UV) light radiation induces the formation of bulky photoproducts in the DNA that globally affect transcription and splicing. However, the signaling pathways and mechanisms that link UV-light-induced DNA damage to changes in RNA metabolism remain poorly understood. Here we employ quantitative phosphoproteomics and protein kinase inhibition to provide a systems view on protein phosphorylation patterns induced by UV light and uncover the dependencies of phosphorylation events on the canonical DNA damage signaling by ATM/ATR and the p38 MAP kinase pathway. We identify RNA-binding proteins as primary substrates and 14-3-3 as direct readers of p38-MK2-dependent phosphorylation induced by UV light. Mechanistically, we show that MK2 phosphorylates the RNA-binding subunit of the NELF complex NELFE on Serine 115. NELFE phosphorylation promotes the recruitment of 14-3-3 and rapid dissociation of the NELF complex from chromatin, which is accompanied by RNA polymerase II elongation.
- Institute of Molecular Biology (IMB), Ackermannweg 4, 55128, Mainz, Germany.
Organizational Affiliation: