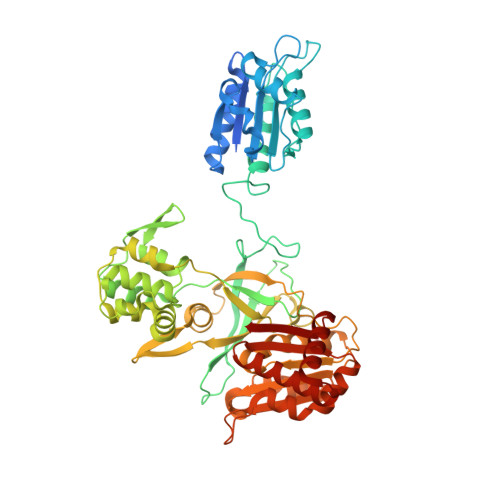
NADPH-dependent sulfite reductase flavoprotein adopts an extended conformation unique to this diflavin reductase.
Tavolieri, A.M., Murray, D.T., Askenasy, I., Pennington, J.M., McGarry, L., Stanley, C.B., Stroupe, M.E.(2019) J Struct Biol 205: 170-179
- PubMed: 30654136
- DOI: https://doi.org/10.1016/j.jsb.2019.01.001
- Primary Citation of Related Structures:
6EFV - PubMed Abstract:
This is the first X-ray crystal structure of the monomeric form of sulfite reductase (SiR) flavoprotein (SiRFP-60) that shows the relationship between its major domains in an extended position not seen before in any homologous diflavin reductases. Small angle neutron scattering confirms this novel domain orientation also occurs in solution. Activity measurements of SiR and SiRFP variants allow us to propose a novel mechanism for electron transfer from the SiRFP reductase subunit to its oxidase metalloenzyme partner that, together, make up the SiR holoenzyme. Specifically, we propose that SiR performs its 6-electron reduction via intramolecular or intermolecular electron transfer. Our model explains both the significance of the stoichiometric mismatch between reductase and oxidase subunits in the holoenzyme and how SiR can handle such a large volume electron reduction reaction that is at the heart of the sulfur bio-geo cycle.
- Department of Biological Science and Institute of Molecular Biophysics, 91 Chieftain Way, Tallahassee, FL 32306, USA.
Organizational Affiliation: