Structure of a RiPP maturase, SkfB
Grell, T.A.J., Kincannon, W.M., Bruender, N.A., Blaesi, E.J., Krebs, C., Bandarian, V., Drennan, C.L.(2018) J Biological Chem
Experimental Data Snapshot
(2018) J Biological Chem
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sporulation killing factor maturation protein SkfB | 412 | Bacillus subtilis subsp. subtilis str. 168 | Mutation(s): 0 Gene Names: skfB, ybcP, ybcQ, BSU01920 EC: 1.21.98 | 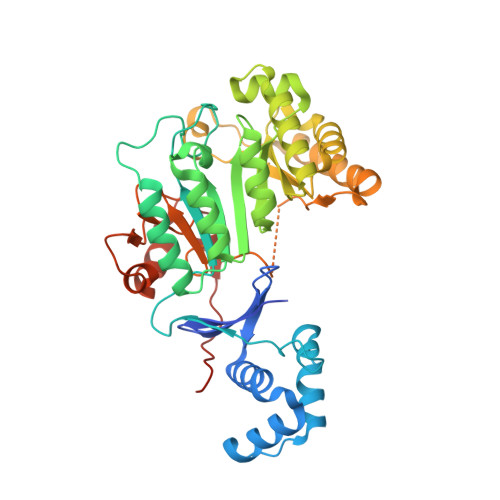 | |
UniProt | |||||
Find proteins for O31423 (Bacillus subtilis (strain 168)) Explore O31423 Go to UniProtKB: O31423 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O31423 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SAM (Subject of Investigation/LOI) Query on SAM | B [auth A] | S-ADENOSYLMETHIONINE C15 H22 N6 O5 S MEFKEPWMEQBLKI-FCKMPRQPSA-N |  | ||
| SF4 (Subject of Investigation/LOI) Query on SF4 | C [auth A] | IRON/SULFUR CLUSTER Fe4 S4 LJBDFODJNLIPKO-UHFFFAOYSA-N |  | ||
| FES (Subject of Investigation/LOI) Query on FES | D [auth A] | FE2/S2 (INORGANIC) CLUSTER Fe2 S2 NIXDOXVAJZFRNF-UHFFFAOYSA-N |  | ||
| MG Query on MG | E [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 76.462 | α = 90 |
| b = 83.11 | β = 94.38 |
| c = 61.299 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| SHELXCD | phasing |
| SHARP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Science Foundation (NSF, United States) | United States | 1122374 |