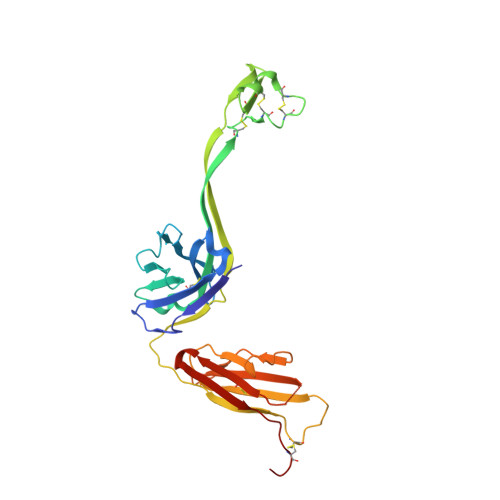
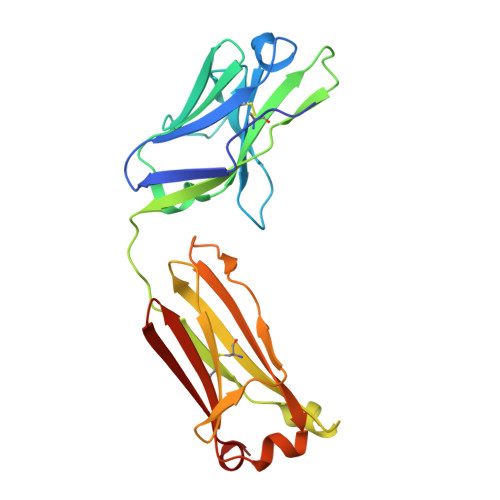
Structural Diversity of Ultralong CDRH3s in Seven Bovine Antibody Heavy Chains.
Dong, J., Finn, J.A., Larsen, P.A., Smith, T.P.L., Crowe Jr., J.E.(2019) Front Immunol 10: 558-558
- PubMed: 30967877
- DOI: https://doi.org/10.3389/fimmu.2019.00558
- Primary Citation of Related Structures:
6E8V, 6E9G, 6E9H, 6E9I, 6E9K, 6E9Q, 6E9U - PubMed Abstract:
Antigen recognition by mammalian antibodies represents the most diverse setting for protein-protein interactions, because antibody variable regions contain exceptionally diverse variable gene repertoires of DNA sequences containing combinatorial, non-templated junctional mutational diversity. Some animals use additional strategies to achieve structural complexity in the antibody combining site, and one of the most interesting of these is the formation of ultralong heavy chain complementarity determining region 3 loops in cattle. Repertoire sequencing studies of bovine antibody heavy chain variable sequences revealed that bovine antibodies can contain heavy chain complementarity determining region 3 (CDRH3) loops with 60 or more amino acids, with complex structures stabilized by multiple disulfide bonds. It is clear that bovine antibodies can achieve long, peculiarly structured CDR3s, but the range of diversity and complexity of those structures is poorly understood. We determined the atomic resolution structure of seven ultralong bovine CDRH3 loops. The studies, combined with five previous structures, reveal a large diversity of cysteine pairing variations, and highly diverse globular domains.
- Vanderbilt Vaccine Center, Vanderbilt University Medical Center, Nashville, TN, United States.
Organizational Affiliation: