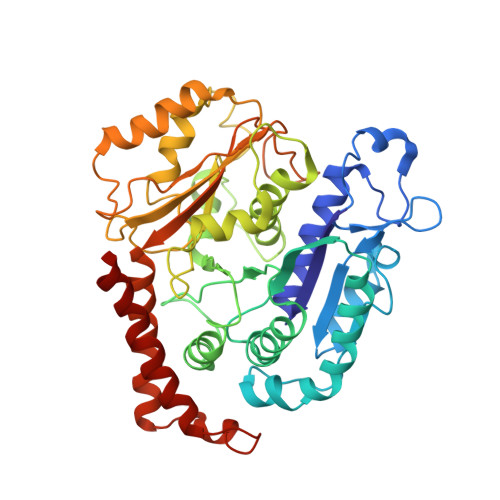
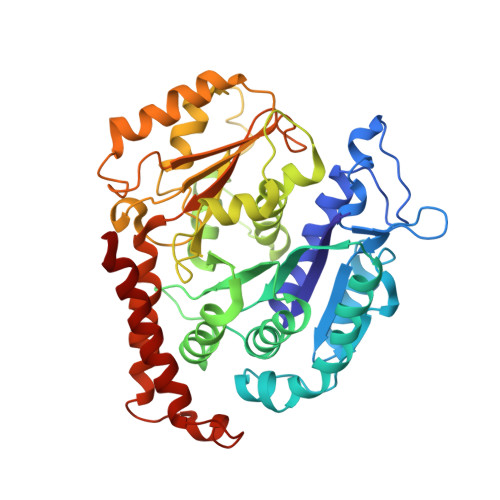
The Structure and Dynamics of C. elegans Tubulin Reveals the Mechanistic Basis of Microtubule Growth.
Chaaban, S., Jariwala, S., Hsu, C.T., Redemann, S., Kollman, J.M., Muller-Reichert, T., Sept, D., Bui, K.H., Brouhard, G.J.(2018) Dev Cell 47: 191
- PubMed: 30245157
- DOI: https://doi.org/10.1016/j.devcel.2018.08.023
- Primary Citation of Related Structures:
6E88 - PubMed Abstract:
The dynamic instability of microtubules is a conserved and fundamental mechanism in eukaryotes. Yet microtubules from different species diverge in their growth rates, lattice structures, and responses to GTP hydrolysis. Therefore, we do not know what limits microtubule growth, what determines microtubule structure, or whether the mechanisms of dynamic instability are universal. Here, we studied microtubules from the nematode C. elegans, which have strikingly fast growth rates and non-canonical lattices in vivo. Using a reconstitution approach, we discovered that C. elegans microtubules combine intrinsically fast growth with very frequent catastrophes. We solved the structure of C. elegans microtubules to 4.8 Å and discovered sequence divergence in the lateral contact loops, one of which is ordered in C. elegans but unresolved in other species. We provide direct evidence that C. elegans tubulin has a higher free energy in solution and propose a model wherein the ordering of lateral contact loops activates tubulin for growth.
- Department of Biology, 1205 Avenue Docteur Penfield, Montréal, QC H3A 1B1, Canada.
Organizational Affiliation: