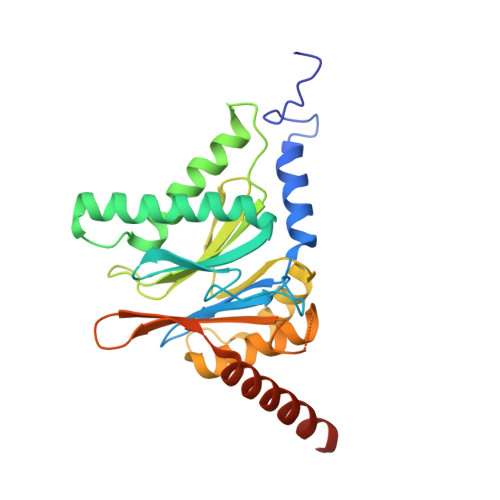
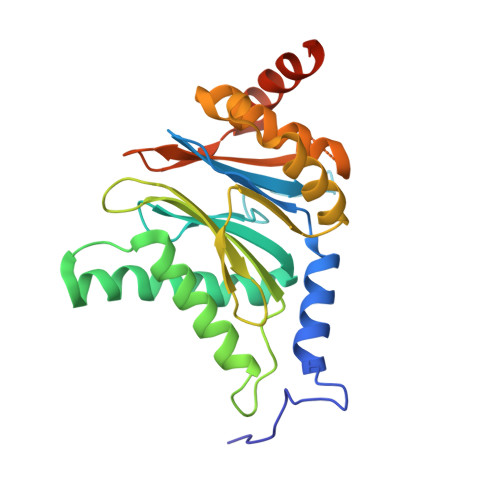
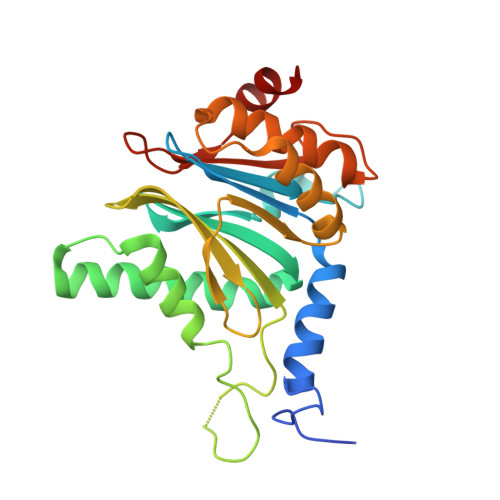
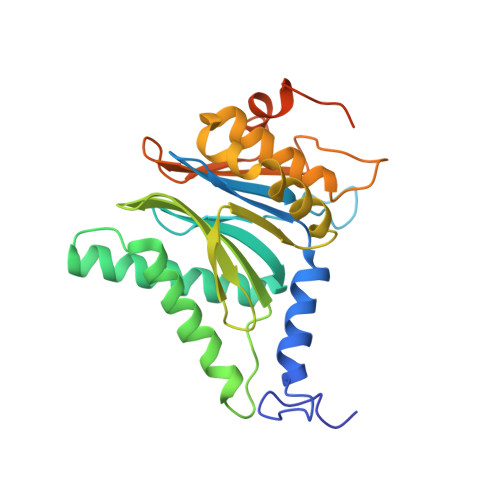
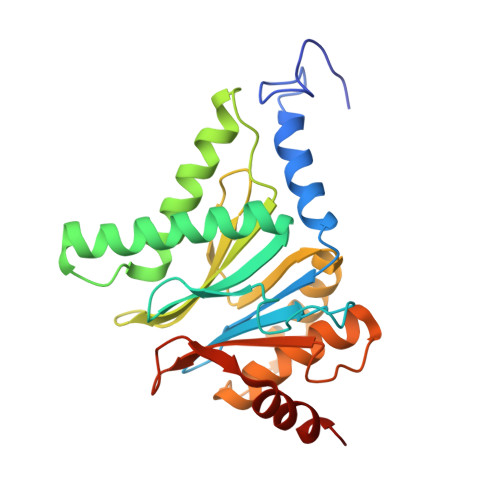
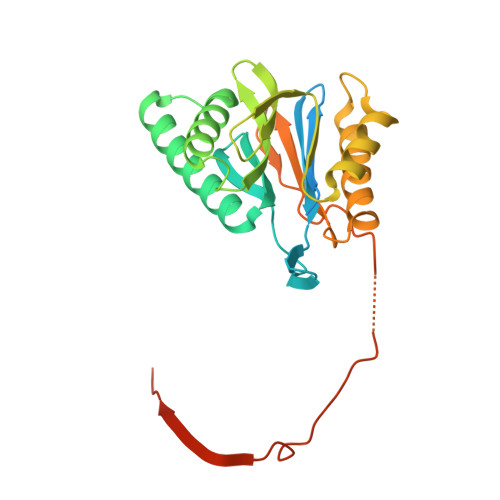
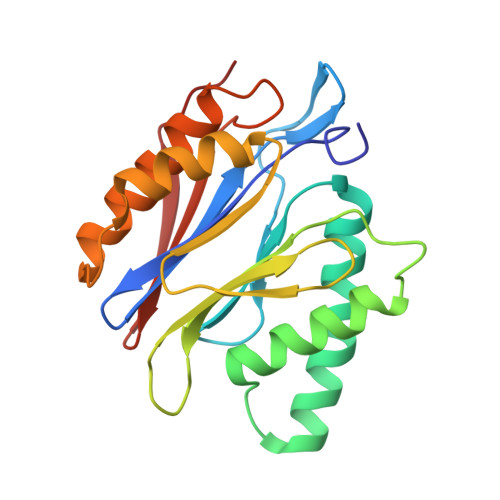
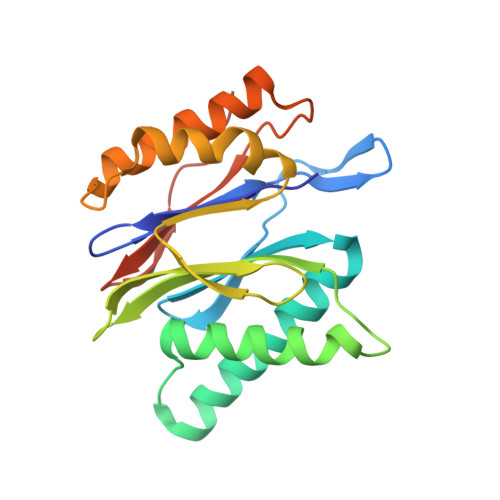
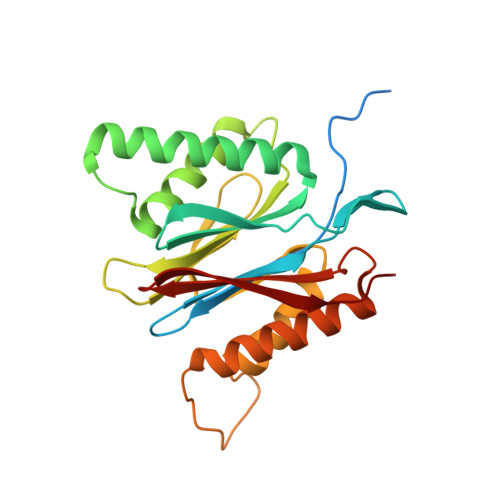
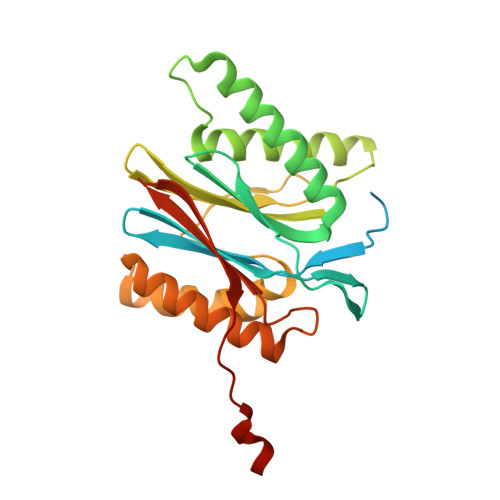
Design and Evaluation of Highly Selective Human Immunoproteasome Inhibitors Reveal a Compensatory Process That Preserves Immune Cell Viability.
Ladi, E., Everett, C., Stivala, C.E., Daniels, B.E., Durk, M.R., Harris, S.F., Huestis, M.P., Purkey, H.E., Staben, S.T., Augustin, M., Blaesse, M., Steinbacher, S., Eidenschenk, C., Pappu, R., Siu, M.(2019) J Med Chem 62: 7032-7041
- PubMed: 31283222
- DOI: https://doi.org/10.1021/acs.jmedchem.9b00509
- Primary Citation of Related Structures:
6E5B - PubMed Abstract:
The pan-proteasome inhibitor bortezomib demonstrated clinical efficacy in off-label trials of Systemic Lupus Erythematosus. One potential mechanism of this clinical benefit is from the depletion of pathogenic immune cells (plasmablasts and plasmacytoid dendritic cells). However, bortezomib is cytotoxic against nonimmune cells, which limits its use for autoimmune diseases. An attractive alternative is to selectively inhibit the immune cell-specific immunoproteasome to deplete pathogenic immune cells and spare nonhematopoietic cells. Here, we disclose the development of highly subunit-selective immunoproteasome inhibitors using insights obtained from the first bona fide human immunoproteasome cocrystal structures. Evaluation of these inhibitors revealed that immunoproteasome-specific inhibition does not lead to immune cell death as anticipated and that targeting viability requires inhibition of both immuno- and constitutive proteasomes. CRISPR/Cas9-mediated knockout experiments confirmed upregulation of the constitutive proteasome upon disruption of the immunoproteasome, protecting cells from death. Thus, immunoproteasome inhibition alone is not a suitable approach to deplete immune cells.
- Proteros Biostructures GmbH , Bunsenstrasse 7a , Planegg-Martinsried 82152 , Germany.
Organizational Affiliation: