Crystal structure of dihydrofolate reductase from Staphylococcus aureus MW2 bound to NADP and p218
Edwards, T.E., Mayclin, S.J., Lorimer, D.D., Horanyi, P.S., Seattle Structural Genomics Center for Infectious Disease (SSGCID)To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Dihydrofolate reductase | 182 | Staphylococcus aureus | Mutation(s): 0 Gene Names: folA EC: 1.5.1.3 | 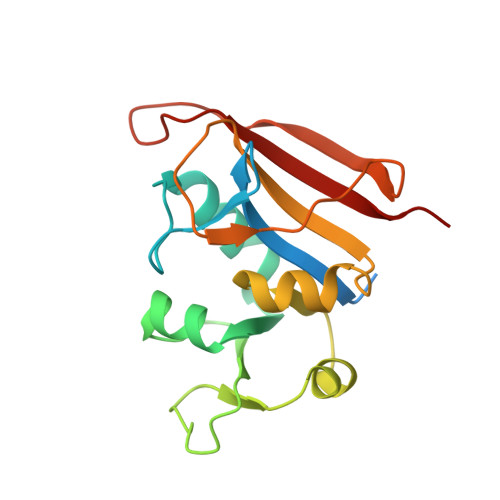 | |
UniProt | |||||
Find proteins for P0A017 (Staphylococcus aureus) Explore P0A017 Go to UniProtKB: P0A017 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A017 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAP Query on NAP | B [auth A] | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE C21 H28 N7 O17 P3 XJLXINKUBYWONI-NNYOXOHSSA-N |  | ||
| MMV Query on MMV | C [auth A] | 3-(2-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)propanoic acid C18 H24 N4 O4 VDGXZSSDCDPCRF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 79 | α = 90 |
| b = 79 | β = 90 |
| c = 108.93 | γ = 120 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |