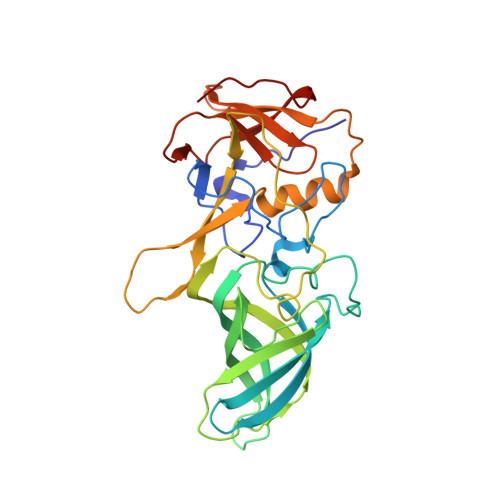
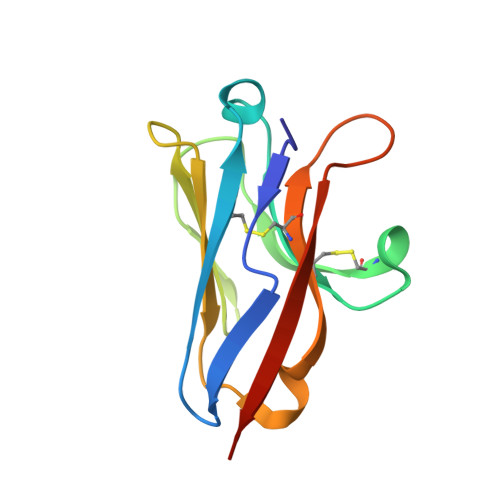
Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor.
Nelson, C.A., Wilen, C.B., Dai, Y.N., Orchard, R.C., Kim, A.S., Stegeman, R.A., Hsieh, L.L., Smith, T.J., Virgin, H.W., Fremont, D.H.(2018) Proc Natl Acad Sci U S A 115: E9201-E9210
- PubMed: 30194229
- DOI: https://doi.org/10.1073/pnas.1805797115
- Primary Citation of Related Structures:
6C6Q, 6C74, 6CRJ, 6E47, 6E48 - PubMed Abstract:
Murine norovirus (MNoV) is closely related to human norovirus (HNoV), an infectious agent responsible for acute gastroenteritis worldwide. Here we report the X-ray crystal structure of the dimeric MNoV VP1 protruding (P) domain in complex with its cellular receptor CD300lf. CD300lf binds the P domain with a 2:2 stoichiometry, engaging a cleft between the AB and DE loops of the P2 subdomain at a site that overlaps the epitopes of neutralizing antibodies. We also identify that bile acids are cofactors enhancing MNoV cell-binding and infectivity. Structures of CD300lf-P domain in complex with glycochenodeoxycholic acid (GCDCA) and lithocholic acid (LCA) reveal two bile acid binding sites at the P domain dimer interface distant from receptor binding sites. The structural determinants for receptor and bile acid binding are supported by numerous biophysical assays utilizing interface residue mutations. We find that the monomeric affinity of CD300lf for the P domain is low and is divalent cation dependent. We have also determined the crystal structure of CD300lf in complex with phosphocholine, revealing that MNoV engages its receptor in a manner mimicking host ligands including similar metal coordination. Docking of the cocomplex structures onto a cryo-EM-derived model of MNoV suggests that each virion can make multiple CD300lf engagements, and thus, infection may be driven by the avidity of cell surface clustered CD300lf. These studies identify multiple potential modulators of norovirus infection that may act to regulate the interaction between the viral capsid P domain and its cognate cellular receptor.
- Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO 63110.
Organizational Affiliation: