Mechanism for the Regulated Control of Bacterial Transcription Termination by a Universal Adaptor Protein.
Lawson, M.R., Ma, W., Bellecourt, M.J., Artsimovitch, I., Martin, A., Landick, R., Schulten, K., Berger, J.M.(2018) Mol Cell 71: 911-922.e4
- PubMed: 30122535
- DOI: https://doi.org/10.1016/j.molcel.2018.07.014
- Primary Citation of Related Structures:
6DUQ - PubMed Abstract:
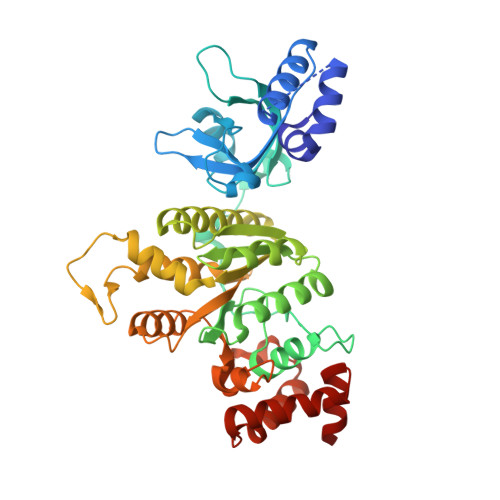
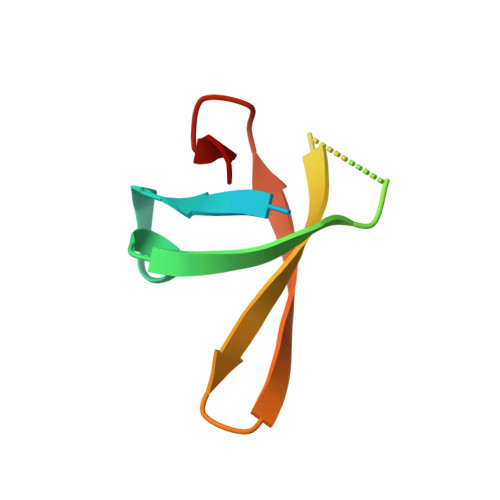
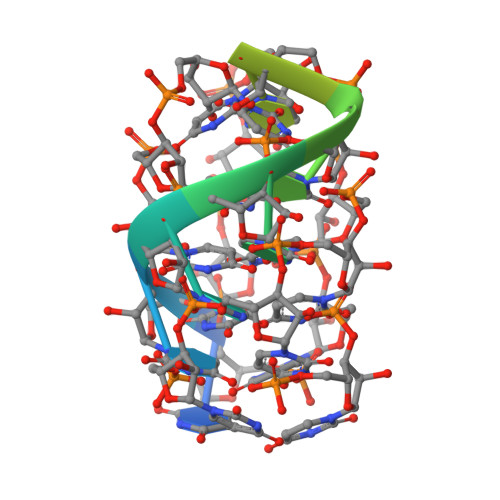
NusG/Spt5 proteins are the only transcription factors utilized by all cellular organisms. In enterobacteria, NusG antagonizes the transcription termination activity of Rho, a hexameric helicase, during the synthesis of ribosomal and actively translated mRNAs. Paradoxically, NusG helps Rho act on untranslated transcripts, including non-canonical antisense RNAs and those arising from translational stress; how NusG fulfills these disparate functions is unknown. Here, we demonstrate that NusG activates Rho by assisting helicase isomerization from an open-ring, RNA-loading state to a closed-ring, catalytically active translocase. A crystal structure of closed-ring Rho in complex with NusG reveals the physical basis for this activation and further explains how Rho is excluded from translationally competent RNAs. This study demonstrates how a universally conserved transcription factor acts to modulate the activity of a ring-shaped ATPase motor and establishes how the innate sequence bias of a termination factor can be modulated to silence pervasive, aberrant transcription.
- Department of Molecular and Cell Biology, University of California at Berkeley, Berkeley, CA 94720, USA.
Organizational Affiliation: