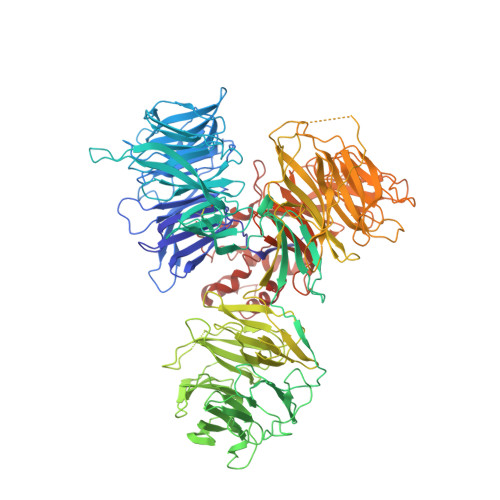
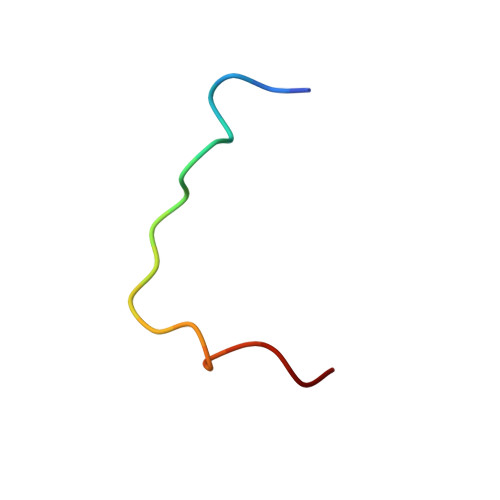
Structural insights into DDA1 function as a core component of the CRL4-DDB1 ubiquitin ligase.
Shabek, N., Ruble, J., Waston, C.J., Garbutt, K.C., Hinds, T.R., Li, T., Zheng, N.(2018) Cell Discov 4: 67-67
- PubMed: 30564455
- DOI: https://doi.org/10.1038/s41421-018-0064-8
- Primary Citation of Related Structures:
6DSZ - 1Department of Pharmacology, University of Washington, Box 357280, Seattle, WA 98195 USA.
Organizational Affiliation: