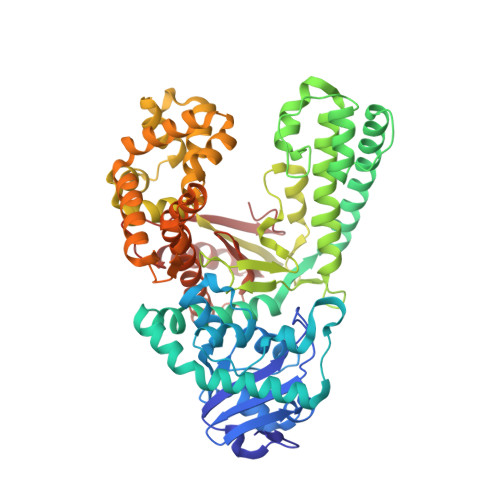
Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Chim, N., Jackson, L.N., Trinh, A.M., Chaput, J.C.(2018) Elife 7
- PubMed: 30338759
- DOI: https://doi.org/10.7554/eLife.40444
- Primary Citation of Related Structures:
6DSU, 6DSV, 6DSW, 6DSX, 6DSY - PubMed Abstract:
High resolution crystal structures of DNA polymerase intermediates are needed to study the mechanism of DNA synthesis in cells. Here we report five crystal structures of DNA polymerase I that capture new conformations for the polymerase translocation and nucleotide pre-insertion steps in the DNA synthesis pathway. We suggest that these new structures, along with previously solved structures, highlight the dynamic nature of the finger subdomain in the enzyme active site.
- Departments of Pharmaceutical Sciences, University of California, Irvine, California.
Organizational Affiliation: