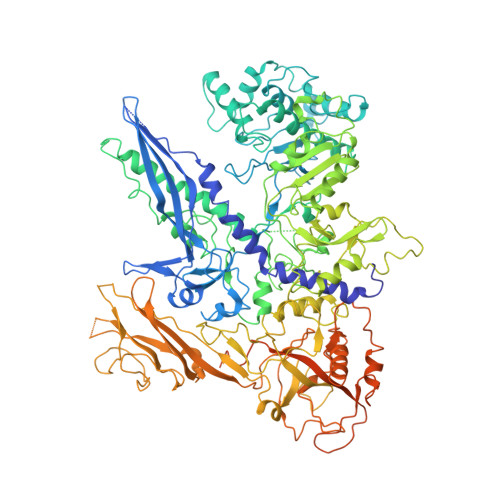
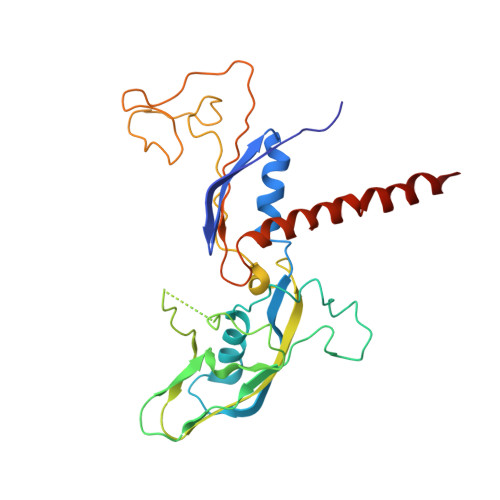
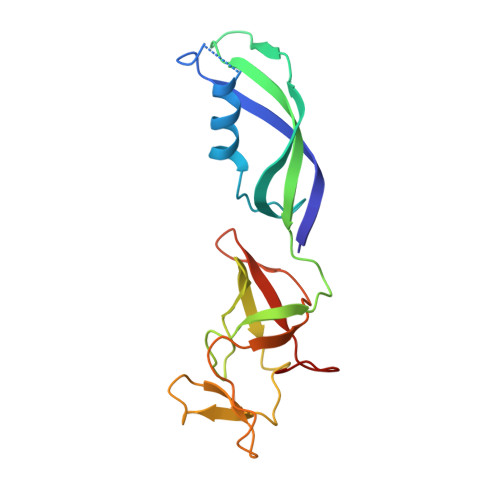
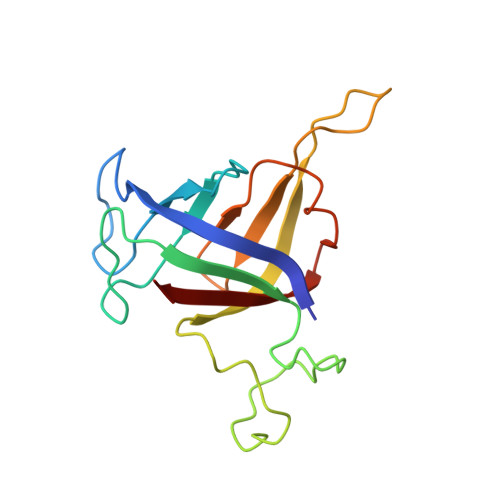
Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1.
Jishage, M., Yu, X., Shi, Y., Ganesan, S.J., Chen, W.Y., Sali, A., Chait, B.T., Asturias, F.J., Roeder, R.G.(2018) Nat Struct Mol Biol 25: 859-867
- PubMed: 30190596
- DOI: https://doi.org/10.1038/s41594-018-0118-5
- Primary Citation of Related Structures:
6DRD, 8ZZP - PubMed Abstract:
Tight binding of Gdown1 represses RNA polymerase II (Pol II) function in a manner that is reversed by Mediator, but the structural basis of these processes is unclear. Although Gdown1 is intrinsically disordered, its Pol II interacting domains were localized and shown to occlude transcription factor IIF (TFIIF) and transcription factor IIB (TFIIB) binding by perfect positioning on their Pol II interaction sites. Robust binding of Gdown1 to Pol II is established by cooperative interactions of a strong Pol II binding region and two weaker binding modulatory regions, thus providing a mechanism both for tight Pol II binding and transcription inhibition and for its reversal. In support of a physiological function for Gdown1 in transcription repression, Gdown1 co-localizes with Pol II in transcriptionally silent nuclei of early Drosophila embryos but re-localizes to the cytoplasm during zygotic genome activation. Our study reveals a self-inactivation through Gdown1 binding as a unique mode of repression in Pol II function.
- Laboratory of Biochemistry and Molecular Biology, The Rockefeller University, New York, NY, USA.
Organizational Affiliation: