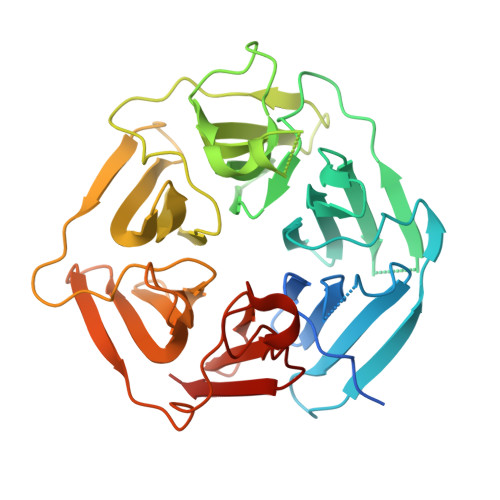
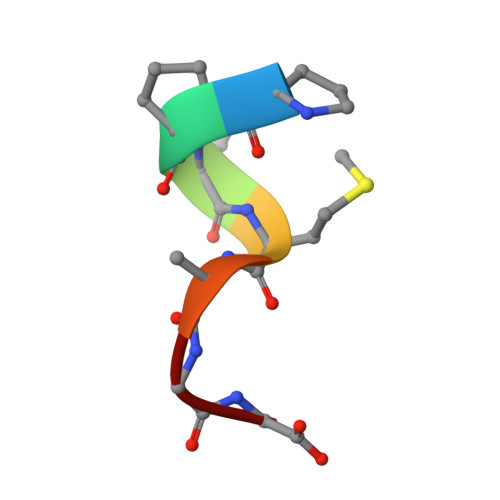
Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Rusnac, D.V., Lin, H.C., Canzani, D., Tien, K.X., Hinds, T.R., Tsue, A.F., Bush, M.F., Yen, H.S., Zheng, N.(2018) Mol Cell 72: 813-822.e4
- PubMed: 30526872
- DOI: https://doi.org/10.1016/j.molcel.2018.10.021
- Primary Citation of Related Structures:
6DO3, 6DO4, 6DO5 - PubMed Abstract:
Aberrant proteins can be deleterious to cells and are cleared by the ubiquitin-proteasome system. A group of C-end degrons that are recognized by specific cullin-RING ubiquitin E3 ligases (CRLs) has recently been identified in some of these abnormal polypeptides. Here, we report three crystal structures of a CRL2 substrate receptor, KLHDC2, in complex with the diglycine-ending C-end degrons of two early-terminated selenoproteins and the N-terminal proteolytic fragment of USP1. The E3 recognizes the degron peptides in a similarly coiled conformation and cradles their C-terminal diglycine with a deep surface pocket. By hydrogen bonding with multiple backbone carbonyls of the peptides, KLHDC2 further locks in the otherwise degenerate degrons with a compact interface and unexpected high affinities. Our results reveal the structural mechanism by which KLHDC2 recognizes the simplest C-end degron and suggest a functional necessity of the E3 to tightly maintain the low abundance of its select substrates.
- Howard Hughes Medical Institute, Department of Pharmacology, University of Washington, Seattle, WA 98195, USA.
Organizational Affiliation: