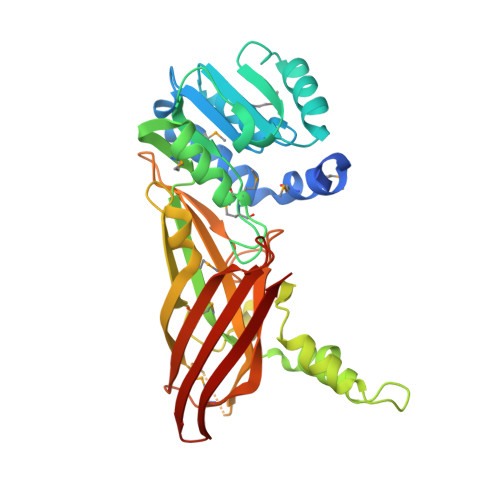
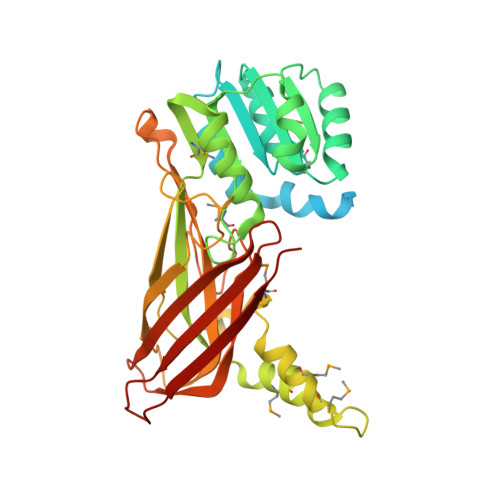
Structural Basis of Protein Arginine Methyltransferase Activation by a Catalytically Dead Homolog (Prozyme).
Hashimoto, H., Kafkova, L., Raczkowski, A., Jordan, K.D., Read, L.K., Debler, E.W.(2020) J Mol Biology 432: 410-426
- PubMed: 31726063
- DOI: https://doi.org/10.1016/j.jmb.2019.11.002
- Primary Citation of Related Structures:
6DNZ - PubMed Abstract:
Prozymes are pseudoenzymes that stimulate the function of weakly active enzymes through complex formation. The major Trypanosoma brucei protein arginine methyltransferase, TbPRMT1 enzyme (ENZ), requires TbPRMT1 prozyme (PRO) to form an active heterotetrameric complex. Here, we present the X-ray crystal structure of the TbPRMT1 ENZ-Δ52PRO tetrameric complex with the cofactor product S-adenosyl-l-homocysteine (AdoHcy) at 2.4 Å resolution. The individual ENZ and PRO units adopt the highly-conserved PRMT domain architecture and form an antiparallel heterodimer that corresponds to the canonical homodimer observed in all previously reported PRMTs. In turn, two such heterodimers assemble into a tetramer both in the crystal and in solution with twofold rotational symmetry. ENZ is unstable in absence of PRO and incapable of forming a homodimer due to a steric clash of an ENZ-specific tyrosine within the dimerization arm, rationalizing why PRO is required to complement ENZ to form a PRMT dimer that is necessary, but not sufficient for PRMT activity. The PRO structure deviates from other, active PRMTs in that it lacks the conserved η2 3 10 -helix within the Rossmann fold, abolishing cofactor binding. In addition to its chaperone function for ENZ, PRO substantially contributes to substrate binding. Heterotetramerization is required for catalysis, as heterodimeric ENZ-PRO mutants lack binding affinity and methyltransferase activity toward the substrate protein TbRGG1. Together, we provide a structural basis for TbPRMT1 ENZ activation by PRO heterotetramer formation, which is conserved across all kinetoplastids, and describe a chaperone function of the TbPRMT1 prozyme, which represents a novel mode of PRMT regulation.
- Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, PA 19107, USA.
Organizational Affiliation: