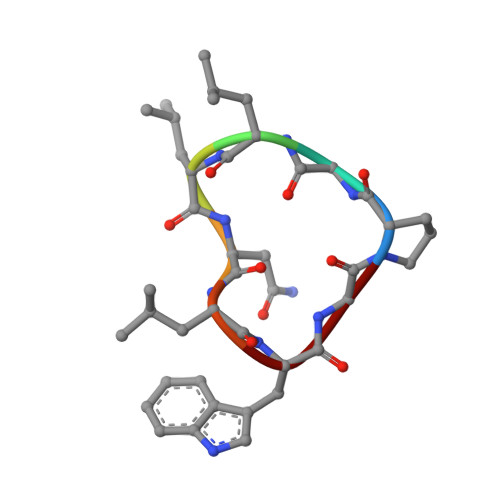
Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
Ramalho, S.D., Wang, C.K., King, G.J., Byriel, K.A., Huang, Y.H., Bolzani, V.S., Craik, D.J.(2018) J Nat Prod 81: 2436-2445
- PubMed: 30345754
- DOI: https://doi.org/10.1021/acs.jnatprod.8b00447
- Primary Citation of Related Structures:
6DKY, 6DKZ, 6DL0, 6DL1 - PubMed Abstract:
Orbitides are small cyclic peptides with a diverse range of therapeutic bioactivities. They are produced by many plant species, including those of the Jatropha genus. Here, the objective was to provide new structural information on orbitides to complement the growing knowledge base on orbitide sequences and activities by focusing on three Jatropha orbitides: ribifolin (1), pohlianin C (7), and jatrophidin (12). To determine three-dimensional structures, racemic crystallography, an emerging structural technique that enables rapid crystallization of biomolecules by combining equal amounts of the two enantiomers, was used. The high-resolution structure of ribifolin (0.99 Å) was elucidated from its racemate and showed it was identical to the structure crystallized from its l-enantiomer only (1.35 Å). Racemic crystallography was also used to elucidate high-resolution structures of pohlianin C (1.20 Å) and jatrophidin (1.03 Å), for which there was difficulty forming crystals without using racemic mixtures. The structures were used to interpret membrane permeability data in PAMPA and a Caco-2 cell assay, showing they had poor permeability. Overall, the results show racemic crystallography can be used to obtain high-resolution structures of orbitides and is useful when enantiopure samples are difficult to crystallize or solution structures from NMR are of low resolution.
- Institute of Chemistry , São Paulo State University-UNESP , Araraquara , São Paulo 14800-060 , Brazil.
Organizational Affiliation: