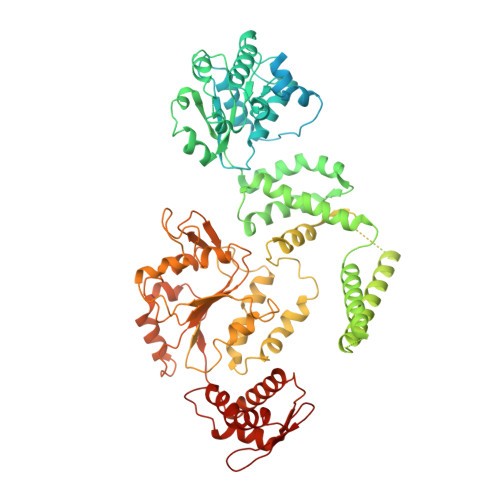
ATP hydrolysis-coupled peptide translocation mechanism ofMycobacterium tuberculosisClpB.
Yu, H., Lupoli, T.J., Kovach, A., Meng, X., Zhao, G., Nathan, C.F., Li, H.(2018) Proc Natl Acad Sci U S A 115: E9560-E9569
- PubMed: 30257943
- DOI: https://doi.org/10.1073/pnas.1810648115
- Primary Citation of Related Structures:
6DJU, 6DJV, 6ED3 - PubMed Abstract:
The protein disaggregase ClpB hexamer is conserved across evolution and has two AAA+-type nucleotide-binding domains, NBD1 and NBD2, in each protomer. In M. tuberculosis ( Mtb ), ClpB facilitates asymmetric distribution of protein aggregates during cell division to help the pathogen survive and persist within the host, but a mechanistic understanding has been lacking. Here we report cryo-EM structures at 3.8- to 3.9-Å resolution of Mtb ClpB bound to a model substrate, casein, in the presence of the weakly hydrolyzable ATP mimic adenosine 5'-[γ-thio]triphosphate. Mtb ClpB existed in solution in two closed-ring conformations, conformers 1 and 2. In both conformers, the 12 pore-loops on the 12 NTDs of the six protomers (P1-P6) were arranged similarly to a staircase around the bound peptide. Conformer 1 is a low-affinity state in which three of the 12 pore-loops (the protomer P1 NBD1 and NBD2 loops and the protomer P2 NBD1 loop) are not engaged with peptide. Conformer 2 is a high-affinity state because only one pore-loop (the protomer P2 NBD1 loop) is not engaged with the peptide. The resolution of the two conformations, along with their bound substrate peptides and nucleotides, enabled us to propose a nucleotide-driven peptide translocation mechanism of a bacterial ClpB that is largely consistent with several recent unfoldase structures, in particular with the eukaryotic Hsp104. However, whereas Hsp104's two NBDs move in opposing directions during one step of peptide translocation, in Mtb ClpB the two NBDs move only in the direction of translocation.
- Cryo-Electron Microscopy Structural Biology Laboratory, Van Andel Research Institute, Grand Rapids, MI 49503.
Organizational Affiliation: