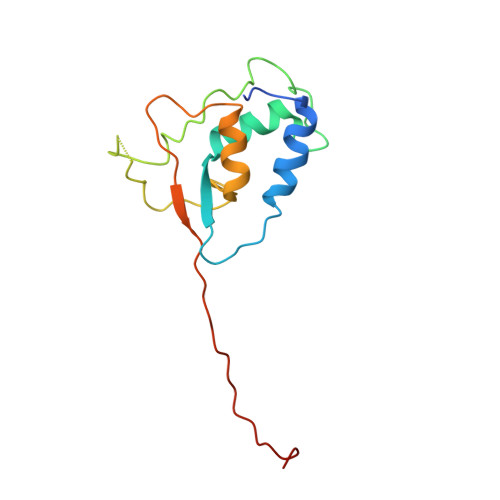
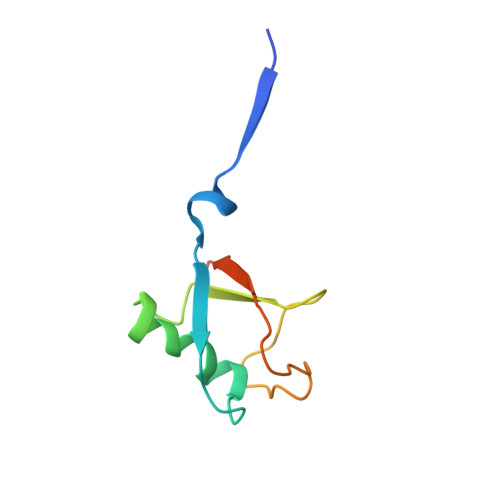
Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Teyra, J., Singer, A.U., Schmitges, F.W., Jaynes, P., Kit Leng Lui, S., Polyak, M.J., Fodil, N., Krieger, J.R., Tong, J., Schwerdtfeger, C., Brasher, B.B., Ceccarelli, D.F.J., Moffat, J., Sicheri, F., Moran, M.F., Gros, P., Eichhorn, P.J.A., Lenter, M., Boehmelt, G., Sidhu, S.S.(2019) Structure 27: 590
- PubMed: 30713027
- DOI: https://doi.org/10.1016/j.str.2019.01.002
- Primary Citation of Related Structures:
6CPM, 6CRN, 6DJ9, 6ML1 - PubMed Abstract:
The multi-domain deubiquitinase USP15 regulates diverse eukaryotic processes and has been implicated in numerous diseases. We developed ubiquitin variants (UbVs) that targeted either the catalytic domain or each of three adaptor domains in USP15, including the N-terminal DUSP domain. We also designed a linear dimer (diUbV), which targeted the DUSP and catalytic domains, and exhibited enhanced specificity and more potent inhibition of catalytic activity than either UbV alone. In cells, the UbVs inhibited the deubiquitination of two USP15 substrates, SMURF2 and TRIM25, and the diUbV inhibited the effects of USP15 on the transforming growth factor β pathway. Structural analyses revealed that three distinct UbVs bound to the catalytic domain and locked the active site in a closed, inactive conformation, and one UbV formed an unusual strand-swapped dimer and bound two DUSP domains simultaneously. These inhibitors will enable the study of USP15 function in oncology, neurology, immunology, and inflammation.
- The Donnelly Centre, University of Toronto, Toronto, ON M5S 3E1, Canada.
Organizational Affiliation: