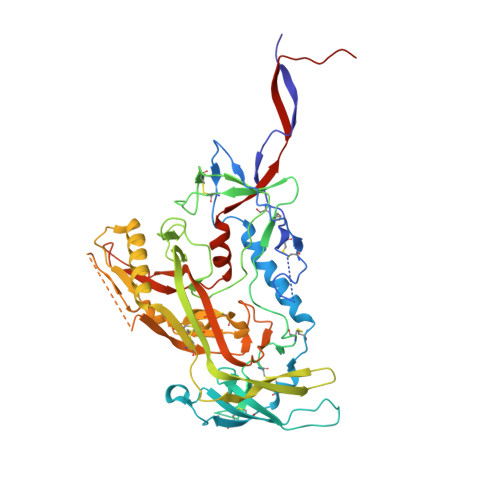
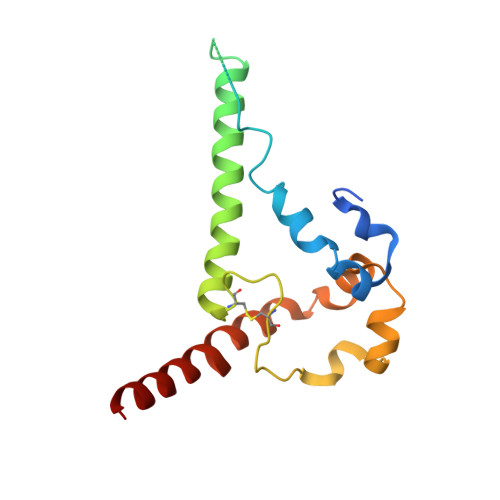
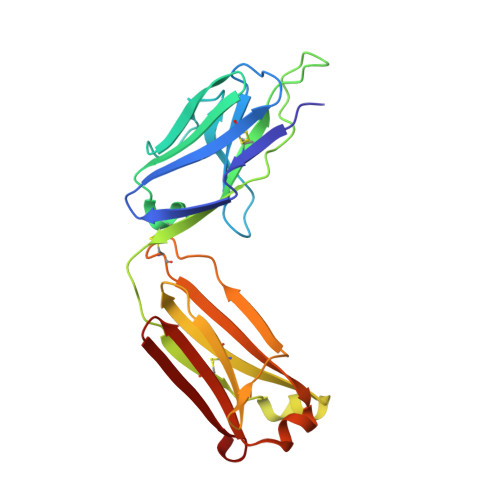
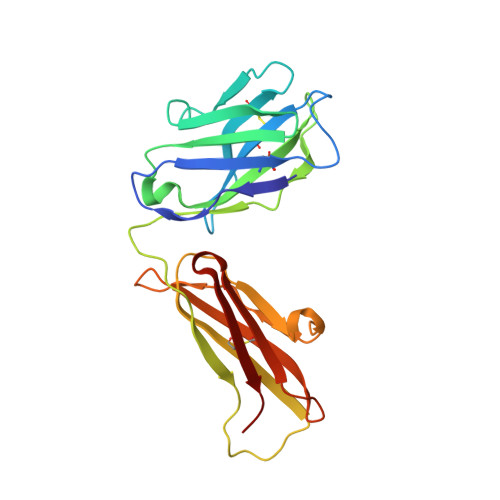
Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization.
Bianchi, M., Turner, H.L., Nogal, B., Cottrell, C.A., Oyen, D., Pauthner, M., Bastidas, R., Nedellec, R., McCoy, L.E., Wilson, I.A., Burton, D.R., Ward, A.B., Hangartner, L.(2018) Immunity 49: 288-300.e8
- PubMed: 30097292
- DOI: https://doi.org/10.1016/j.immuni.2018.07.009
- Primary Citation of Related Structures:
6CJK, 6DID - PubMed Abstract:
Characterizing polyclonal antibody responses via currently available methods is inherently complex and difficult. Mapping epitopes in an immune response is typically incomplete, which creates a barrier to fully understanding the humoral response to antigens and hinders rational vaccine design efforts. Here, we describe a method of characterizing polyclonal responses by using electron microscopy, and we applied this method to the immunization of rabbits with an HIV-1 envelope glycoprotein vaccine candidate, BG505 SOSIP.664. We detected known epitopes within the polyclonal sera and revealed how antibody responses evolved during the prime-boosting strategy to ultimately result in a neutralizing antibody response. We uncovered previously unidentified epitopes, including an epitope proximal to one recognized by human broadly neutralizing antibodies as well as potentially distracting non-neutralizing epitopes. Our method provides an efficient and semiquantitative map of epitopes that are targeted in a polyclonal antibody response and should be of widespread utility in vaccine and infection studies.
- Department of Immunology and Microbiology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: