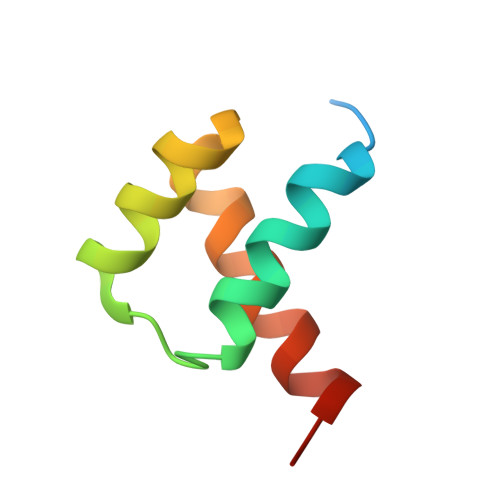
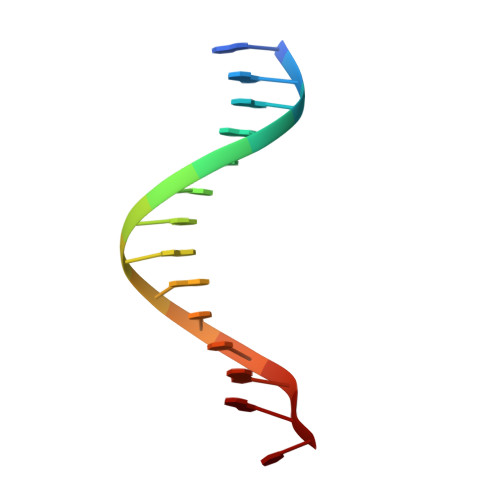
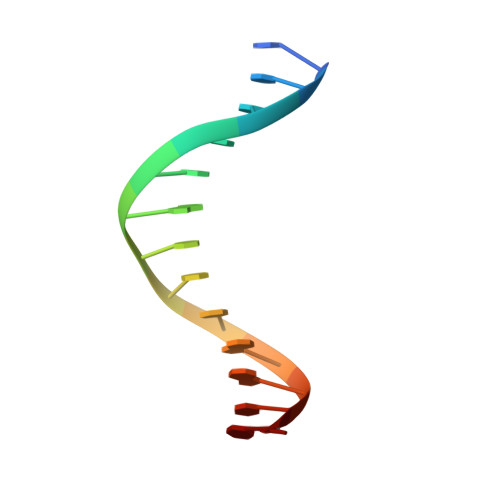
Comment on structural basis of DUX4/IGH-driven transactivation.
Aihara, H., Shi, K., Lee, J.K., Bosnakovski, D., Kyba, M.(2018) Leukemia 32: 2090-2092
- PubMed: 30054554
- DOI: https://doi.org/10.1038/s41375-018-0217-7
- Primary Citation of Related Structures:
6DFY - Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, Minneapolis, MN, 55455, USA. aihar001@umn.edu.
Organizational Affiliation: