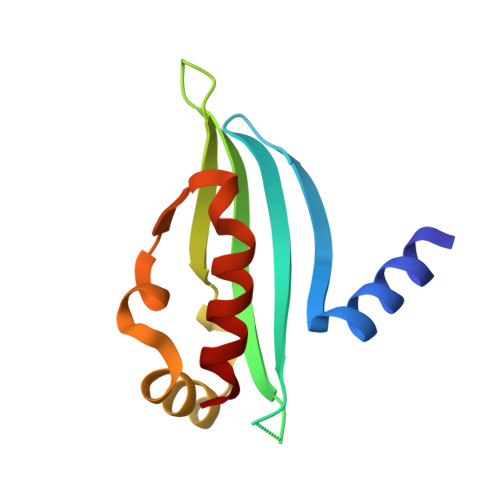
The budding-yeast RWD protein Csm1 scaffolds diverse protein complexes through a conserved structural mechanism.
Singh, N., Corbett, K.D.(2018) Protein Sci 27: 2094-2100
- PubMed: 30252178
- DOI: https://doi.org/10.1002/pro.3515
- Primary Citation of Related Structures:
6DEI - PubMed Abstract:
RWD domains mediate protein-protein interactions in a variety of pathways in eukaryotes. In budding yeast, the RWD domain protein Csm1 is particularly versatile, assembling key complexes in the nucleolus and at meiotic kinetochores through multiple protein interaction surfaces. Here, we reveal a third functional context for Csm1 by identifying a new Csm1-interacting protein, Dse3. We show that Dse3 interacts with Csm1 in a structurally equivalent manner to its known binding partners Mam1 and Ulp2, despite these three proteins' lack of overall sequence homology. We theorize that the unique "clamp" structure of Csm1 and the loose sequence requirements for Csm1 binding have led to its incorporation into at least three different structural/signaling pathways in budding yeast.
- Ludwig Institute for Cancer Research, San Diego Branch, La Jolla, California, 92093.
Organizational Affiliation: