Structural characterization of monoclonal antibodies targeting C-terminal Ser404region of phosphorylated tau protein.
Chukwu, J.E., Congdon, E.E., Sigurdsson, E.M., Kong, X.P.(2019) MAbs 11: 477-488
- PubMed: 30794086
- DOI: https://doi.org/10.1080/19420862.2019.1574530
- Primary Citation of Related Structures:
6DC7, 6DC8, 6DC9, 6DCA - PubMed Abstract:
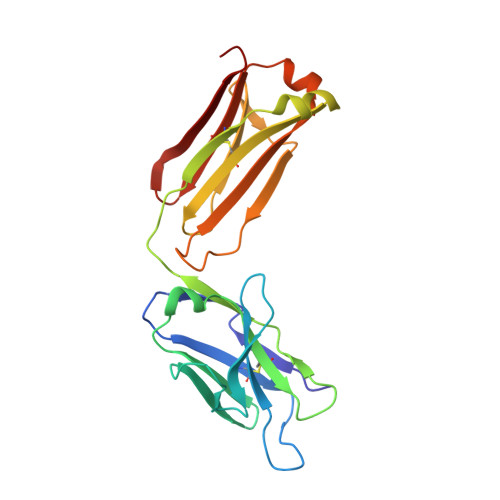
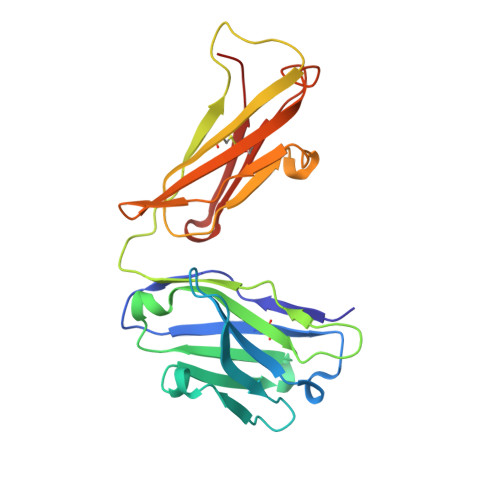
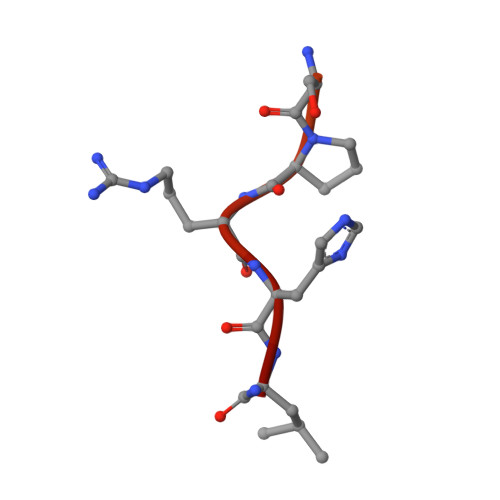
Targeting tau with immunotherapies is currently the most common approach taken in clinical trials of patients with Alzheimer's disease. The most prominent pathological feature of tau is its hyperphosphorylation, which may cause the protein to aggregate into toxic assemblies that collectively lead to neurodegeneration. Of the phospho-epitopes, the region around Ser 396 /Ser 404 has received particular attention for therapeutic targeting because of its prominence and stability in diseased tissue. Herein, we present the antigen-binding fragment (Fab)/epitope complex structures of three different monoclonal antibodies (mAbs) that target the pSer 404 tau epitope region. Most notably, these structures reveal an antigen conformation similar to a previously described pathogenic tau epitope, pSer 422 , which was shown to have a β-strand structure that may be linked to the seeding core in tau oligomers. In addition, we have previously reported on the similarly ordered conformation observed in a pSer 396 epitope, which is in tandem with pSer 404 . Our data are the first Fab structures of mAbs bound to this epitope region of the tau protein and support the existence of proteopathic tau conformations stabilized by specific phosphorylation events that are viable targets for immune modulation.
- a Department of Biochemistry & Molecular Pharmacology , New York University School of Medicine , New York , NY , USA.
Organizational Affiliation: