Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif.
Shelke, S.A., Shao, Y., Laski, A., Koirala, D., Weissman, B.P., Fuller, J.R., Tan, X., Constantin, T.P., Waggoner, A.S., Bruchez, M.P., Armitage, B.A., Piccirilli, J.A.(2018) Nat Commun 9: 4542-4542
- PubMed: 30382099
- DOI: https://doi.org/10.1038/s41467-018-06942-3
- Primary Citation of Related Structures:
6DB8, 6DB9 - PubMed Abstract:
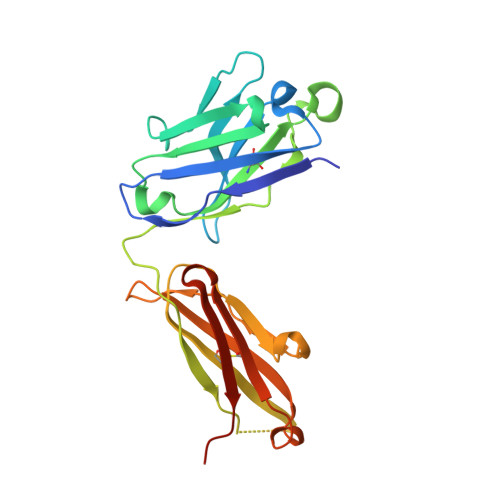
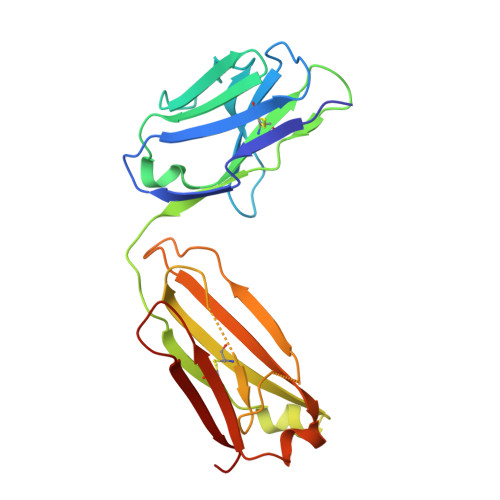
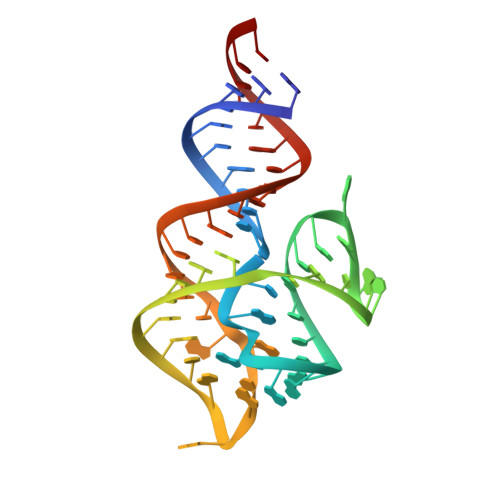
The DIR2s RNA aptamer, a second-generation, in-vitro selected binder to dimethylindole red (DIR), activates the fluorescence of cyanine dyes, DIR and oxazole thiazole blue (OTB), allowing detection of two well-resolved emission colors. Using Fab BL3-6 and its cognate hairpin as a crystallization module, we solved the crystal structures of both the apo and OTB-SO 3 bound forms of DIR2s at 2.0 Å and 1.8 Å resolution, respectively. DIR2s adopts a compact, tuning fork-like architecture comprised of a helix and two short stem-loops oriented in parallel to create the ligand binding site through tertiary interactions. The OTB-SO 3 fluorophore binds in a planar conformation to a claw-like structure formed by a purine base-triple, which provides a stacking platform for OTB-SO 3, and an unpaired nucleotide, which partially caps the binding site from the top. The absence of a G-quartet or base tetrad makes the DIR2s aptamer unique among fluorogenic RNAs with known 3D structure.
- Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL, 60637, USA.
Organizational Affiliation: