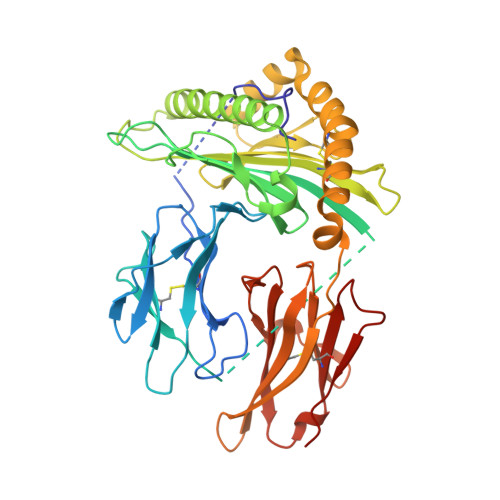
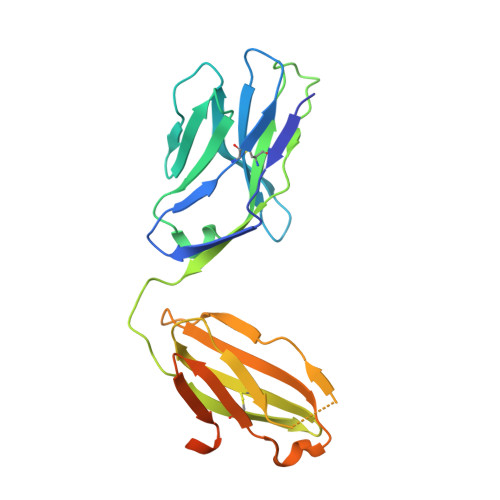
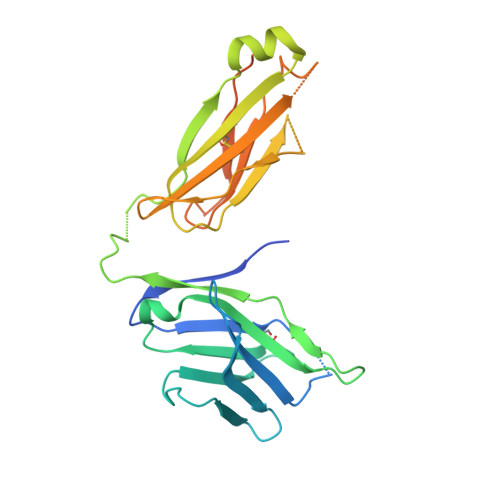
Generation and molecular recognition of melanoma-associated antigen-specific human gamma delta T cells.
Benveniste, P.M., Roy, S., Nakatsugawa, M., Chen, E.L.Y., Nguyen, L., Millar, D.G., Ohashi, P.S., Hirano, N., Adams, E.J., Zuniga-Pflucker, J.C.(2018) Sci Immunol 3
- PubMed: 30552102
- DOI: https://doi.org/10.1126/sciimmunol.aav4036
- Primary Citation of Related Structures:
6D7G - PubMed Abstract:
Antigen recognition by T cells bearing αβ T cell receptors (TCRs) is restricted by major histocompatibility complex (MHC). However, how antigens are recognized by T cells bearing γδ TCRs remains unclear. Although γδ T cells can recognize nonclassical MHC, it is generally thought that recognition of antigens is not MHC restricted. Here, we took advantage of an in vitro system to generate antigen-specific human T cells and show that melanoma-associated antigens, MART-1 and gp100, can be recognized by γδ T cells in an MHC-restricted fashion. Cloning and transferring of MART-1-specific γδ TCRs restored the specific recognition of the initial antigen MHC/peptide reactivity and conferred antigen-specific functional responses. A crystal structure of a MART-1-specific γδ TCR, together with MHC/peptide, revealed distinctive but similar docking properties to those previously reported for αβ TCRs, recognizing MART-1 on HLA-A*0201. Our work shows that antigen-specific and MHC-restricted γδ T cells can be generated in vitro and that MART-1-specific γδ T cells can also be found and cloned from the naïve repertoire. These findings reveal that classical MHC-restricted human γδ TCRs exist in the periphery and have the potential to be used in developing of new TCR-based immunotherapeutic approaches.
- Sunnybrook Research Institute, Toronto, ON, Canada.
Organizational Affiliation: