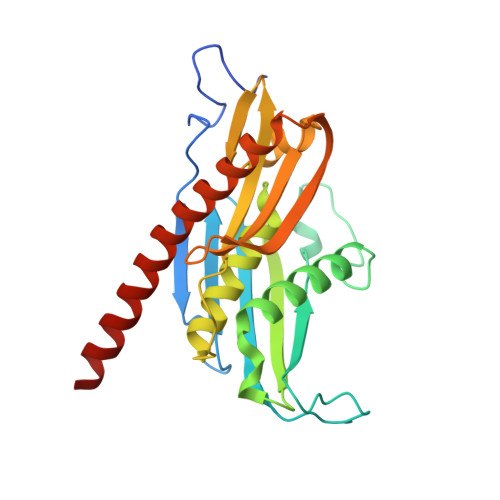
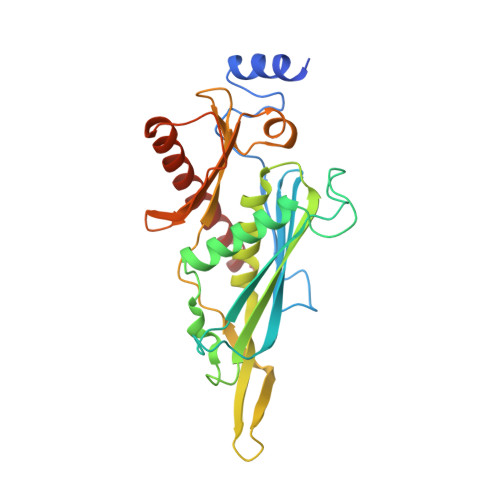
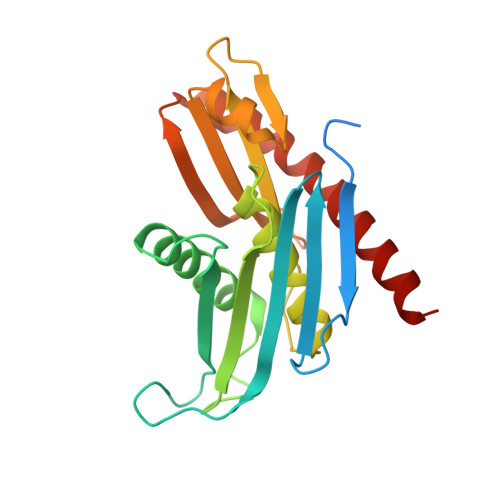
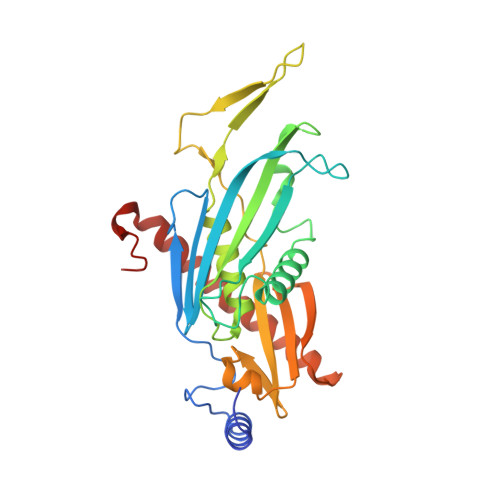
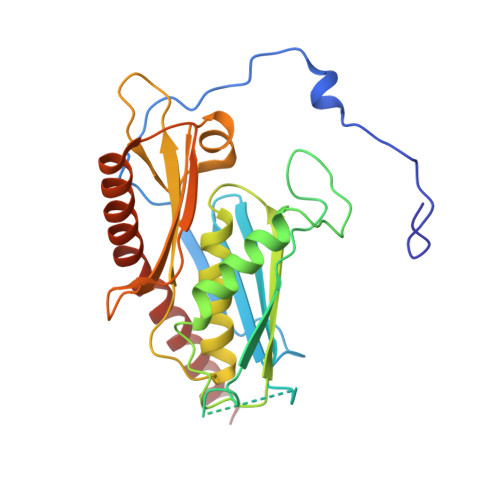
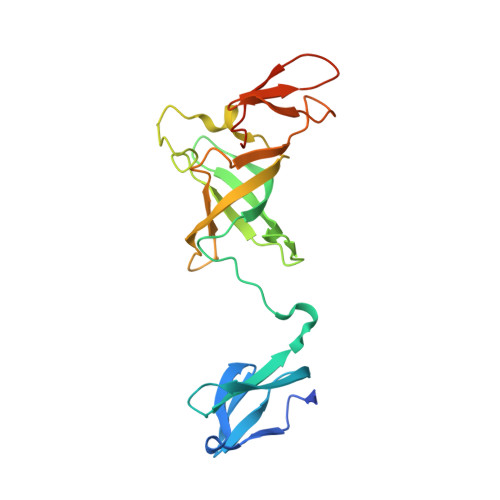
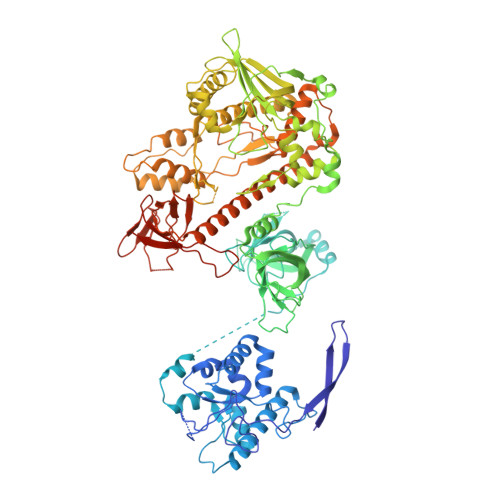
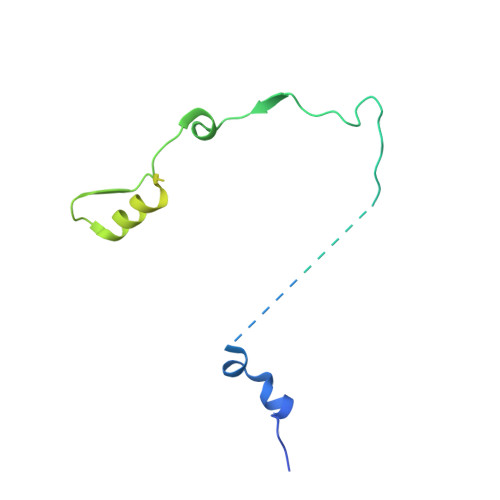
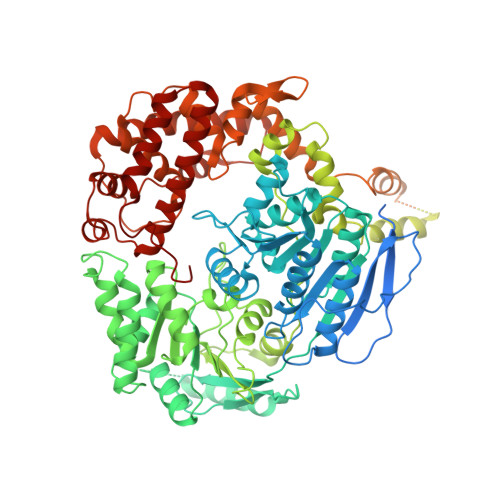
Helicase-Dependent RNA Decay Illuminated by a Cryo-EM Structure of a Human Nuclear RNA Exosome-MTR4 Complex.
Weick, E.M., Puno, M.R., Januszyk, K., Zinder, J.C., DiMattia, M.A., Lima, C.D.(2018) Cell 173: 1663
- PubMed: 29906447
- DOI: https://doi.org/10.1016/j.cell.2018.05.041
- Primary Citation of Related Structures:
6D6Q, 6D6R - PubMed Abstract:
The ribonucleolytic RNA exosome interacts with RNA helicases to degrade RNA. To understand how the 3' to 5' Mtr4 helicase engages RNA and the nuclear exosome, we reconstituted 14-subunit Mtr4-containing RNA exosomes from Saccharomyces cerevisiae, Schizosaccharomyces pombe, and human and show that they unwind structured substrates to promote degradation. We loaded a human exosome with an optimized DNA-RNA chimera that stalls MTR4 during unwinding and determined its structure to an overall resolution of 3.45 Å by cryoelectron microscopy (cryo-EM). The structure reveals an RNA-engaged helicase atop the non-catalytic core, with RNA captured within the central channel and DIS3 exoribonuclease active site. MPP6 tethers MTR4 to the exosome through contacts to the RecA domains of MTR4. EXOSC10 remains bound to the core, but its catalytic module and cofactor C1D are displaced by RNA-engaged MTR4. Competition for the exosome core may ensure that RNA is committed to degradation by DIS3 when engaged by MTR4.
- Structural Biology Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA.
Organizational Affiliation: